Experimental determination of the evolvability of a transcription factor
- PMID: 19841254
- PMCID: PMC2765923
- DOI: 10.1073/pnas.0907688106
Experimental determination of the evolvability of a transcription factor
Abstract
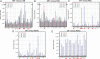
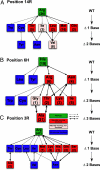
Sequence-specific binding of a transcription factor to DNA is the central event in any transcriptional regulatory network. However, relatively little is known about the evolutionary plasticity of transcription factors. For example, the exact functional consequence of an amino acid substitution on the DNA-binding specificity of most transcription factors is currently not predictable. Furthermore, although the major structural families of transcription factors have been identified, the detailed DNA-binding repertoires within most families have not been characterized. We studied the sequence recognition code and evolvability of the basic helix-loop-helix transcription factor family by creating all possible 95 single-point mutations of five DNA-contacting residues of Max, a human helix-loop-helix transcription factor and measured the detailed DNA-binding repertoire of each mutant. Our results show that the sequence-specific repertoire of Max accessible through single-point mutations is extremely limited, and we are able to predict 92% of the naturally occurring diversity at these positions. All naturally occurring basic regions were also found to be accessible through functional intermediates. Finally, we observed a set of amino acids that are functional in vitro but are not found to be used naturally, indicating that functionality alone is not sufficient for selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

