Identification of SMARCAL1 as a component of the DNA damage response
- PMID: 19841479
- PMCID: PMC2791023
- DOI: 10.1074/jbc.M109.048330
Identification of SMARCAL1 as a component of the DNA damage response
Abstract
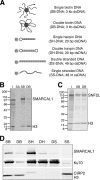
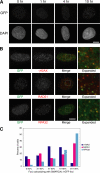
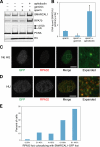
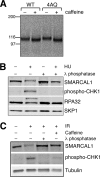
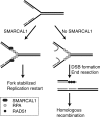
SMARCAL1 (also known as HARP) is a SWI/SNF family protein with an ATPase activity stimulated by DNA containing both single-stranded and double-stranded regions. Mutations in SMARCAL1 are associated with the disease Schimke immuno-osseous dysplasia, a multisystem autosomal recessive disorder characterized by T cell immunodeficiency, growth inhibition, and renal dysfunction. The cellular function of SMARCAL1, however, is unknown. Here, using Xenopus egg extracts and mass spectrometry, we identify SMARCAL1 as a protein recruited to double-stranded DNA breaks. SMARCAL1 binds to double-stranded breaks and stalled replication forks in both egg extract and human cells, specifically colocalizing with the single-stranded DNA binding factor RPA. In addition, SMARCAL1 interacts physically with RPA independently of DNA. SMARCAL1 is phosphorylated in a caffeine-sensitive manner in response to double-stranded breaks and stalled replication forks. It has been suggested that stalled forks can be stabilized by a mechanism involving caffeine-sensitive kinases, or they collapse and subsequently recruit Rad51 to promote homologous recombination repair. We show that depletion of SMARCAL1 from U2OS cells leads to increased frequency of RAD51 foci upon generation of stalled replication forks, indicating that fork breakdown is more prevalent in the absence of SMARCAL1. We propose that SMARCAL1 is a novel DNA damage-binding protein involved in replication fork stabilization.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

