The role of dynamic conformational ensembles in biomolecular recognition
- PMID: 19841628
- PMCID: PMC2916928
- DOI: 10.1038/nchembio.232
The role of dynamic conformational ensembles in biomolecular recognition
Erratum in
- Nat Chem Biol. 2009 Dec;5(12):954
Abstract
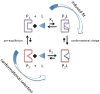
Molecular recognition is central to all biological processes. For the past 50 years, Koshland's 'induced fit' hypothesis has been the textbook explanation for molecular recognition events. However, recent experimental evidence supports an alternative mechanism. 'Conformational selection' postulates that all protein conformations pre-exist, and the ligand selects the most favored conformation. Following binding the ensemble undergoes a population shift, redistributing the conformational states. Both conformational selection and induced fit appear to play roles. Following binding by a primary conformational selection event, optimization of side chain and backbone interactions is likely to proceed by an induced fit mechanism. Conformational selection has been observed for protein-ligand, protein-protein, protein-DNA, protein-RNA and RNA-ligand interactions. These data support a new molecular recognition paradigm for processes as diverse as signaling, catalysis, gene regulation and protein aggregation in disease, which has the potential to significantly impact our views and strategies in drug design, biomolecular engineering and molecular evolution.
Figures

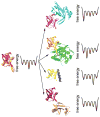
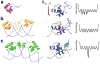
References
-
- Fischer E. Einfluss der configuration auf die Wirkung der Enzyme. Ber Dtsch Chem Ges. 1894;27:2984–2993.
-
- Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–603. - PubMed
-
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

