Marine-derived metabolites of S-adenosylmethionine as templates for new anti-infectives
- PMID: 19841722
- PMCID: PMC2763108
- DOI: 10.3390/md7030401
Marine-derived metabolites of S-adenosylmethionine as templates for new anti-infectives
Abstract
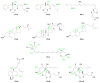
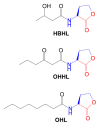
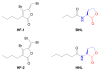
S-Adenosylmethionine (AdoMet) is a key biochemical co-factor whose proximate metabolites include methylated macromolecules (e.g., nucleic acids, proteins, phospholipids), methylated small molecules (e.g., sterols, biogenic amines), polyamines (e.g., spermidine, spermine), ethylene, and N-acyl-homoserine lactones. Marine organisms produce numerous AdoMet metabolites whose novel structures can be regarded as lead compounds for anti-infective drug design.
Keywords: adenosylmethionine; ethylene; polyamines; quorum sensing; radical SAM.
Figures
















References
-
- Kotb M, Geller AM. Methionine Adenosyltransferase-Structure and Function. Pharmacol Ther. 1993;59:125–143. - PubMed
-
- Iwig DF, Booker SJ. Insight into the Polar Reactivity of the Onium Chalcogen Analogues of S-Adenosyl-l-Methionine. Biochemistry. 2004;43:13496–13509. - PubMed
-
- Fontecave M, Atta M, Mulliez E. S-Adenosylmethionine: Nothing Goes to Waste. Trends Biochem Sci. 2004;29:243–249. - PubMed
-
- Kagan RM, Clarke S. Widespread Occurrence of 3 Sequence Motifs in Diverse S-Adenosylmethionine-Dependent Methyltransferases Suggests a Common Structure for These Enzymes. Arch Biochem Biophys. 1994;310:417–427. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
