Differentiation driven changes in the dynamic organization of Basal transcription initiation
- PMID: 19841728
- PMCID: PMC2754661
- DOI: 10.1371/journal.pbio.1000220
Differentiation driven changes in the dynamic organization of Basal transcription initiation
Abstract
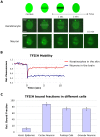
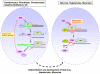
Studies based on cell-free systems and on in vitro-cultured living cells support the concept that many cellular processes, such as transcription initiation, are highly dynamic: individual proteins stochastically bind to their substrates and disassemble after reaction completion. This dynamic nature allows quick adaptation of transcription to changing conditions. However, it is unknown to what extent this dynamic transcription organization holds for postmitotic cells embedded in mammalian tissue. To allow analysis of transcription initiation dynamics directly into living mammalian tissues, we created a knock-in mouse model expressing fluorescently tagged TFIIH. Surprisingly and in contrast to what has been observed in cultured and proliferating cells, postmitotic murine cells embedded in their tissue exhibit a strong and long-lasting transcription-dependent immobilization of TFIIH. This immobilization is both differentiation driven and development dependent. Furthermore, although very statically bound, TFIIH can be remobilized to respond to new transcriptional needs. This divergent spatiotemporal transcriptional organization in different cells of the soma revisits the generally accepted highly dynamic concept of the kinetic framework of transcription and shows how basic processes, such as transcription, can be organized in a fundamentally different fashion in intact organisms as previously deduced from in vitro studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Giglia-Mari G, Coin F, Ranish J. A, Hoogstraten D, Theil A, et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36:714–719. - PubMed
-
- Egly J. M. The 14th Datta Lecture. TFIIH: from transcription to clinic. FEBS Lett. 2001;498:124–128. - PubMed
-
- Mitchell J. R, Hoeijmakers J. H, Niedernhofer L. J. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr Opin Cell Biol. 2003;15:232–240. - PubMed
-
- Hoeijmakers J. H, Egly J. M, Vermeulen W. TFIIH: a key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;6:26–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

