Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication
- PMID: 19841738
- PMCID: PMC2759517
- DOI: 10.1371/journal.pone.0007517
Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication
Abstract
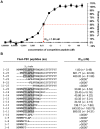
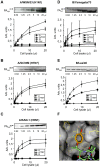
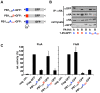
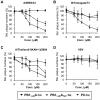
There is an urgent need for new drugs against influenza type A and B viruses due to incomplete protection by vaccines and the emergence of resistance to current antivirals. The influenza virus polymerase complex, consisting of the PB1, PB2 and PA subunits, represents a promising target for the development of new drugs. We have previously demonstrated the feasibility of targeting the protein-protein interaction domain between the PB1 and PA subunits of the polymerase complex of influenza A virus using a small peptide derived from the PA-binding domain of PB1. However, this influenza A virus-derived peptide did not affect influenza B virus polymerase activity. Here we report that the PA-binding domain of the polymerase subunit PB1 of influenza A and B viruses is highly conserved and that mutual amino acid exchange shows that they cannot be functionally exchanged with each other. Based on phylogenetic analysis and a novel biochemical ELISA-based screening approach, we were able to identify an influenza A-derived peptide with a single influenza B-specific amino acid substitution which efficiently binds to PA of both virus types. This dual-binding peptide blocked the viral polymerase activity and growth of both virus types. Our findings provide proof of principle that protein-protein interaction inhibitors can be generated against influenza A and B viruses. Furthermore, this dual-binding peptide, combined with our novel screening method, is a promising platform to identify new antiviral lead compounds.
Conflict of interest statement
Figures


Similar articles
-
Limited compatibility of polymerase subunit interactions in influenza A and B viruses.J Biol Chem. 2010 May 28;285(22):16704-12. doi: 10.1074/jbc.M110.102533. Epub 2010 Apr 2. J Biol Chem. 2010. PMID: 20363752 Free PMC article.
-
Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase.Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6247-52. doi: 10.1073/pnas.1119817109. Epub 2012 Apr 2. Proc Natl Acad Sci U S A. 2012. PMID: 22474359 Free PMC article.
-
Identification of high-affinity PB1-derived peptides with enhanced affinity to the PA protein of influenza A virus polymerase.Antimicrob Agents Chemother. 2011 Feb;55(2):696-702. doi: 10.1128/AAC.01419-10. Epub 2010 Dec 6. Antimicrob Agents Chemother. 2011. PMID: 21135188 Free PMC article.
-
Focusing on the Influenza Virus Polymerase Complex: Recent Progress in Drug Discovery and Assay Development.Curr Med Chem. 2019;26(13):2243-2263. doi: 10.2174/0929867325666180706112940. Curr Med Chem. 2019. PMID: 29984646 Free PMC article. Review.
-
Influenza A virus polymerase: an attractive target for next-generation anti-influenza therapeutics.Drug Discov Today. 2018 Mar;23(3):503-518. doi: 10.1016/j.drudis.2018.01.028. Epub 2018 Jan 12. Drug Discov Today. 2018. PMID: 29339107 Review.
Cited by
-
Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting.PLoS Pathog. 2011 Sep;7(9):e1002187. doi: 10.1371/journal.ppat.1002187. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909257 Free PMC article.
-
Current progress in antiviral strategies.Trends Pharmacol Sci. 2014 Feb;35(2):86-102. doi: 10.1016/j.tips.2013.11.006. Epub 2014 Jan 14. Trends Pharmacol Sci. 2014. PMID: 24439476 Free PMC article. Review.
-
Antiviral strategies against influenza virus: towards new therapeutic approaches.Cell Mol Life Sci. 2014 Oct;71(19):3659-83. doi: 10.1007/s00018-014-1615-2. Epub 2014 Apr 4. Cell Mol Life Sci. 2014. PMID: 24699705 Free PMC article. Review.
-
Crucial role of PA in virus life cycle and host adaptation of influenza A virus.Med Microbiol Immunol. 2015 Apr;204(2):137-49. doi: 10.1007/s00430-014-0349-y. Epub 2014 Jul 29. Med Microbiol Immunol. 2015. PMID: 25070354 Review.
-
Mouse adaptation of influenza B virus increases replication in the upper respiratory tract and results in droplet transmissibility in ferrets.Sci Rep. 2015 Nov 3;5:15940. doi: 10.1038/srep15940. Sci Rep. 2015. PMID: 26526113 Free PMC article.
References
-
- Tumpey TM, Belser JA. Resurrected Pandemic Influenza Viruses. Annu Rev Microbiol 2009 - PubMed
-
- Parrish CR, Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol. 2005;59:553–586. - PubMed
-
- Baine WB, Luby JP, Martin SM. Severe illness with influenza B. Am J Med. 1980;68:181–189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

