Probing neuroserpin polymerization and interaction with amyloid-beta peptides using single molecule fluorescence
- PMID: 19843463
- PMCID: PMC2764104
- DOI: 10.1016/j.bpj.2009.07.057
Probing neuroserpin polymerization and interaction with amyloid-beta peptides using single molecule fluorescence
Abstract
Neuroserpin is a member of the serine proteinase inhibitor superfamily. It can undergo a conformational transition to form polymers that are associated with the dementia familial encephalopathy with neuroserpin inclusion bodies and the wild-type protein can inhibit the toxicity of amyloid-beta peptides in Alzheimer's disease. We have used a single molecule fluorescence method, two color coincidence detection, to determine the rate-limiting steps of the early stages of the polymerization of fluorophore-labeled neuroserpin and have assessed how this process is altered in the presence of A beta(1-40.) Our data show that neuroserpin polymerization proceeds first by the unimolecular formation of an active monomer, followed by competing processes of both polymerization and formation of a latent monomer from the activated species. These data are not in keeping with the recently proposed domain swap model of polymer formation in which the latent species and activated monomer are likely to be formed by competing pathways directly from the unactivated monomeric serpin. Moreover, the A beta(1-40) peptide forms a weak complex with neuroserpin (dissociation constant of 10 +/- 5 nM) that increases the amount of active monomer thereby increasing the rate of polymerization. The A beta(1-40) is displaced from the complex so that it acts as a catalyst and is not incorporated into neuroserpin polymers.
Figures




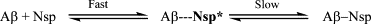
Similar articles
-
The tempered polymerization of human neuroserpin.PLoS One. 2012;7(3):e32444. doi: 10.1371/journal.pone.0032444. Epub 2012 Mar 6. PLoS One. 2012. PMID: 22412873 Free PMC article.
-
Mutant Neuroserpin (S49P) that causes familial encephalopathy with neuroserpin inclusion bodies is a poor proteinase inhibitor and readily forms polymers in vitro.J Biol Chem. 2002 May 10;277(19):17367-73. doi: 10.1074/jbc.M200680200. Epub 2002 Mar 5. J Biol Chem. 2002. PMID: 11880376
-
pH-dependent stability of neuroserpin is mediated by histidines 119 and 138; implications for the control of beta-sheet A and polymerization.Protein Sci. 2010 Feb;19(2):220-8. doi: 10.1002/pro.299. Protein Sci. 2010. PMID: 19953505 Free PMC article.
-
Neuroserpin: a serpin to think about.Cell Mol Life Sci. 2006 Mar;63(6):709-22. doi: 10.1007/s00018-005-5077-4. Cell Mol Life Sci. 2006. PMID: 16465451 Free PMC article. Review.
-
Neuroserpin.Front Biosci. 2006 Jan 1;11:33-45. doi: 10.2741/1778. Front Biosci. 2006. PMID: 16146712 Review.
Cited by
-
Glaucoma is associated with plasmin proteolytic activation mediated through oxidative inactivation of neuroserpin.Sci Rep. 2017 Aug 21;7(1):8412. doi: 10.1038/s41598-017-08688-2. Sci Rep. 2017. PMID: 28827627 Free PMC article.
-
Functional and dysfunctional conformers of human neuroserpin characterized by optical spectroscopies and Molecular Dynamics.Biochim Biophys Acta. 2015 Feb;1854(2):110-7. doi: 10.1016/j.bbapap.2014.10.002. Epub 2014 Nov 6. Biochim Biophys Acta. 2015. PMID: 25450507 Free PMC article.
-
Two latent and two hyperstable polymeric forms of human neuroserpin.Biophys J. 2010 Nov 17;99(10):3402-11. doi: 10.1016/j.bpj.2010.09.021. Biophys J. 2010. PMID: 21081089 Free PMC article.
-
Probing the folding pathway of a consensus serpin using single tryptophan mutants.Sci Rep. 2018 Feb 1;8(1):2121. doi: 10.1038/s41598-018-19567-9. Sci Rep. 2018. PMID: 29391487 Free PMC article.
-
The probable role of tissue plasminogen activator/neuroserpin axis in Alzheimer's disease: a new perspective.Acta Neurol Belg. 2024 Apr;124(2):377-388. doi: 10.1007/s13760-023-02403-x. Epub 2023 Nov 2. Acta Neurol Belg. 2024. PMID: 37917293 Free PMC article. Review.
References
-
- Selkoe D.J. Alzheimer's disease: genes, proteins, and therapy. Physiol. Rev. 2001;81:741–766. - PubMed
-
- Kinghorn K.J., Crowther D.C., Sharp L.K., Nerelius C., Davis R.L. Neuroserpin binds Aβ and is a neuroprotective component of amyloid plaques in Alzheimer disease. J. Biol. Chem. 2006;281:29268–29277. - PubMed
-
- Silverman G.A., Bird P.I., Carrell R.W., Church F.C., Coughlin P.B. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, novel functions, mechanism of inhibition and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. - PubMed
-
- Galliciotti G., Sonderegger P. Neuroserpin. Front. Biosci. 2006;11:33–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

