Common structural transitions in explicit-solvent simulations of villin headpiece folding
- PMID: 19843466
- PMCID: PMC2764099
- DOI: 10.1016/j.bpj.2009.08.012
Common structural transitions in explicit-solvent simulations of villin headpiece folding
Abstract
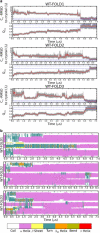
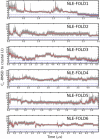
Molecular dynamics simulations of protein folding can provide very high-resolution data on the folding process; however, due to computational challenges most studies of protein folding have been limited to small peptides, or made use of approximations such as Gō potentials or implicit solvent models. We have performed a set of molecular dynamics simulations totaling >50 micros on the villin headpiece subdomain, one of the most stable and fastest-folding naturally occurring proteins, in explicit solvent. We find that the wild-type villin headpiece reliably folds to a native conformation on timescales similar to experimentally observed folding, but that a fast folding double-norleucine mutant shows significantly more heterogeneous behavior. Along with other recent simulation studies, we note the occurrence of nonnative structures intermediates, which may yield a nativelike signal in the fluorescence measurements typically used to study villin folding. Based on the wild-type simulations, we propose alternative approaches to measure the formation of the native state.
Figures

References
-
- Yang W.Y., Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. - PubMed
-
- Kubelka J., Hofrichter J., Eaton W.A. The protein folding ‘speed limit’. Curr. Opin. Struct. Biol. 2004;14:76–88. - PubMed
-
- Kubelka J., Eaton W.A., Hofrichter J. Experimental tests of villin subdomain folding simulations. J. Mol. Biol. 2003;329:625–630. - PubMed
-
- Kubelka J., Chiu T.K., Davies D.R., Eaton W.A., Hofrichter J. Sub-microsecond protein folding. J. Mol. Biol. 2006;359:546–553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

