Live cell analysis of aquaporin-4 m1/m23 interactions and regulated orthogonal array assembly in glial cells
- PMID: 19843522
- PMCID: PMC2791014
- DOI: 10.1074/jbc.M109.071670
Live cell analysis of aquaporin-4 m1/m23 interactions and regulated orthogonal array assembly in glial cells
Abstract
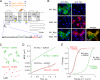
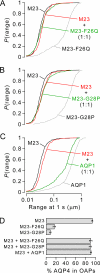
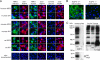
Aquaporin-4 (AQP4) can assemble into supramolecular aggregates called orthogonal arrays of particles (OAPs). In cells expressing single AQP4 isoforms, we found previously that OAP formation by AQP4-M23 requires N terminus interactions just downstream of Met-23 and that the inability of AQP4-M1 to form OAPs involves blocking by residues upstream of Met-23. Here, we studied M1/M23 interactions and regulated OAP assembly by nanometer-resolution tracking of quantum dot-labeled AQP4 in live cells expressing differentially tagged AQP4 isoforms and in primary glial cell cultures in which native AQP4 was labeled with a monoclonal recombinant neuromyelitis optica autoantibody. OAP assembly was assessed independently by Blue Native gel electrophoresis. We found that OAPs in native glial cells could be reproduced in transfected cells expressing equal amounts of AQP4-M1 and -M23. Mutants of M23 that do not themselves form OAPs, including M23-F26Q and M23-G28P, were able to fully co-associate with native M23 to form large immobile OAPs. Analysis of a palmitoylation-null M1 mutant (C13A/C17A) indicated palmitoylation-dependent OAP assembly only in the presence of M23, with increased M1 palmitoylation causing progressive OAP disruption. Differential regulation of OAP assembly by palmitoylation, calcium elevation, and protein kinase C activation was found in primary glial cell cultures. We conclude that M1 and M23 co-assemble in AQP4 OAPs and that specific signaling events can regulate OAP assembly in glial cells.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

