Proteomics analysis reveals overlapping functions of clustered protocadherins
- PMID: 19843561
- PMCID: PMC2808268
- DOI: 10.1074/mcp.M900343-MCP200
Proteomics analysis reveals overlapping functions of clustered protocadherins
Abstract
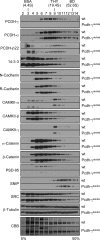
The three tandem-arrayed protocadherin (Pcdh) gene clusters, namely Pcdh-alpha, Pcdh-beta, and Pcdh-gamma, play important roles in the development of the vertebrate central nervous system. To gain insight into the molecular action of PCDHs, we performed a systematic proteomics analysis of PCDH-gamma-associated protein complexes. We identified a list of 154 non-redundant proteins in the PCDH-gamma complexes. This list includes nearly 30 members of clustered Pcdh-alpha, -beta, and -gamma families as core components of the complexes and additionally over 120 putative PCDH-associated proteins. We validated a selected subset of PCDH-gamma-associated proteins using specific antibodies. Analysis of the identities of PCDH-associated proteins showed that the majority of them overlap with the proteomic profile of postsynaptic density preparations. Further analysis of membrane protein complexes revealed that several validated PCDH-gamma-associated proteins exhibit reduced levels in Pcdh-gamma-deficient brain tissues. Therefore, PCDH-gamma s are required for the integrity of the complexes. However, the size of the overall complexes and the abundance of many other proteins remained unchanged, raising a possibility that PCDH-alphas and PCDH-betas might compensate for PCDH-gamma function in complex formation. As a test of this idea, RNA interference knockdown of both PCDH-alphas and PCDH-gamma s showed that PCDHs have redundant functions in regulating neuronal survival in the chicken spinal cord. Taken together, our data provide evidence that clustered PCDHs coexist in large protein complexes and have overlapping functions during vertebrate neural development.
Figures






References
-
- Benson D. L., Colman D. R., Huntley G. W. (2001) Molecules, maps and synapse specificity. Nat. Rev. Neurosci. 2, 899–909 - PubMed
-
- Takeichi M. (2007) The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20 - PubMed
-
- Morishita H., Yagi T. (2007) Protocadherin family: diversity, structure, and function. Curr. Opin. Cell Biol. 19, 584–592 - PubMed
-
- Halbleib J. M., Nelson W. J. (2006) Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20, 3199–3214 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

