A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide
- PMID: 19843957
- PMCID: PMC2778577
- DOI: 10.1126/scisignal.2000599
A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide
Abstract
Nearly identical cells can exhibit substantially different responses to the same stimulus. We monitored the nuclear localization dynamics of nuclear factor kappaB (NF-kappaB) in single cells stimulated with tumor necrosis factor-alpha (TNF-alpha) and lipopolysaccharide (LPS). Cells stimulated with TNF-alpha have quantitative differences in NF-kappaB nuclear localization, whereas LPS-stimulated cells can be clustered into transient or persistent responders, representing two qualitatively different groups based on the NF-kappaB response. These distinct behaviors can be linked to a secondary paracrine signal secreted at low concentrations, such that not all cells undergo a second round of NF-kappaB activation. From our single-cell data, we built a computational model that captures cell variability, as well as population behaviors. Our findings show that mammalian cells can create "noisy" environments to produce diversified responses to stimuli.
Figures
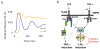




References
-
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–50. - PubMed
-
- Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–9. - PubMed
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

