Arabidopsis gene co-expression network and its functional modules
- PMID: 19845953
- PMCID: PMC2772859
- DOI: 10.1186/1471-2105-10-346
Arabidopsis gene co-expression network and its functional modules
Abstract
Background: Biological networks characterize the interactions of biomolecules at a systems-level. One important property of biological networks is the modular structure, in which nodes are densely connected with each other, but between which there are only sparse connections. In this report, we attempted to find the relationship between the network topology and formation of modular structure by comparing gene co-expression networks with random networks. The organization of gene functional modules was also investigated.
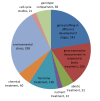
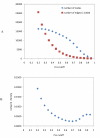
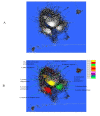
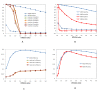
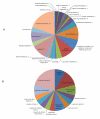
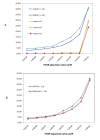
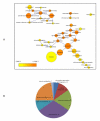
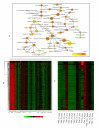
Results: We constructed a genome-wide Arabidopsis gene co-expression network (AGCN) by using 1094 microarrays. We then analyzed the topological properties of AGCN and partitioned the network into modules by using an efficient graph clustering algorithm. In the AGCN, 382 hub genes formed a clique, and they were densely connected only to a small subset of the network. At the module level, the network clustering results provide a systems-level understanding of the gene modules that coordinate multiple biological processes to carry out specific biological functions. For instance, the photosynthesis module in AGCN involves a very large number (> 1000) of genes which participate in various biological processes including photosynthesis, electron transport, pigment metabolism, chloroplast organization and biogenesis, cofactor metabolism, protein biosynthesis, and vitamin metabolism. The cell cycle module orchestrated the coordinated expression of hundreds of genes involved in cell cycle, DNA metabolism, and cytoskeleton organization and biogenesis. We also compared the AGCN constructed in this study with a graphical Gaussian model (GGM) based Arabidopsis gene network. The photosynthesis, protein biosynthesis, and cell cycle modules identified from the GGM network had much smaller module sizes compared with the modules found in the AGCN, respectively.
Conclusion: This study reveals new insight into the topological properties of biological networks. The preferential hub-hub connections might be necessary for the formation of modular structure in gene co-expression networks. The study also reveals new insight into the organization of gene functional modules.
Figures


References
-
- Freeman TC, Goldovsky L, Brosch M, Van Dongen S, Maziere P, Grocock RJ, Freilich S, Thornton J, Enright AJ. Construction, visualisation, and clustering of transcription networks from Microarray expression data. PLoS Comput Biol. 2007;3:2032–2042. doi: 10.1371/journal.pcbi.0030206. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

