Genomic and epigenetic evidence for oxytocin receptor deficiency in autism
- PMID: 19845972
- PMCID: PMC2774338
- DOI: 10.1186/1741-7015-7-62
Genomic and epigenetic evidence for oxytocin receptor deficiency in autism
Abstract
Background: Autism comprises a spectrum of behavioral and cognitive disturbances of childhood development and is known to be highly heritable. Although numerous approaches have been used to identify genes implicated in the development of autism, less than 10% of autism cases have been attributed to single gene disorders.
Methods: We describe the use of high-resolution genome-wide tilepath microarrays and comparative genomic hybridization to identify copy number variants within 119 probands from multiplex autism families. We next carried out DNA methylation analysis by bisulfite sequencing in a proband and his family, expanding this analysis to methylation analysis of peripheral blood and temporal cortex DNA of autism cases and matched controls from independent datasets. We also assessed oxytocin receptor (OXTR) gene expression within the temporal cortex tissue by quantitative real-time polymerase chain reaction (PCR).
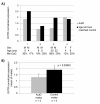
Results: Our analysis revealed a genomic deletion containing the oxytocin receptor gene, OXTR (MIM accession no.: 167055), previously implicated in autism, was present in an autism proband and his mother who exhibits symptoms of obsessive-compulsive disorder. The proband's affected sibling did not harbor this deletion but instead may exhibit epigenetic misregulation of this gene through aberrant gene silencing by DNA methylation. Further DNA methylation analysis of the CpG island known to regulate OXTR expression identified several CpG dinucleotides that show independent statistically significant increases in the DNA methylation status in the peripheral blood cells and temporal cortex in independent datasets of individuals with autism as compared to control samples. Associated with the increase in methylation of these CpG dinucleotides is our finding that OXTR mRNA showed decreased expression in the temporal cortex tissue of autism cases matched for age and sex compared to controls.
Conclusion: Together, these data provide further evidence for the role of OXTR and the oxytocin signaling pathway in the etiology of autism and, for the first time, implicate the epigenetic regulation of OXTR in the development of the disorder.See the related commentary by Gurrieri and Neri: http://www.biomedcentral.com/1741-7015/7/63.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

