Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids
- PMID: 19846551
- PMCID: PMC2790983
- DOI: 10.1074/jbc.M109.072132
Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids
Abstract
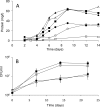
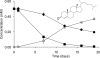
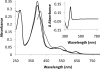
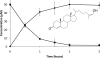
Cyp125 (Rv3545c), a cytochrome P450, is encoded as part of the cholesterol degradation gene cluster conserved among members of the Mycobacterium tuberculosis complex. This enzyme has been implicated in mycobacterial pathogenesis, and a homologue initiates cholesterol catabolism in the soil actinomycete Rhodococcus jostii RHA1. In Mycobacterium bovis BCG, cyp125 was up-regulated 7.1-fold with growth on cholesterol. A cyp125 deletion mutant of BCG did not grow on cholesterol and accumulated 4-cholesten-3-one when incubated in the presence of cholesterol. Wild-type BCG grew on this metabolite. By contrast, a parallel cyp125 deletion mutation of M. tuberculosis H37Rv did not affect growth on cholesterol. Purified Cyp125 from M. tuberculosis, heterologously produced in R. jostii RHA1, bound cholesterol and 4-cholesten-3-one with apparent dissociation constants of 0.20 +/- 0.02 microM and 0.27 +/- 0.05 microm, respectively. When reconstituted with KshB, the cognate reductase of the ketosteroid 9alpha-hydroxylase, Cyp125 catalyzed the hydroxylation of these steroids. MS and NMR analyses revealed that hydroxylation occurred at carbon 26 of the steroid side chain, allowing unambiguous classification of Cyp125 as a steroid C26-hydroxylase. This study establishes the catalytic function of Cyp125 and, in identifying an important difference in the catabolic potential of M. bovis and M. tuberculosis, suggests that Cyp125 may have an additional function in pathogenesis.
Figures



References
-
- World Health Organization (2008) Anti-tuberculosis Drug Resistance in the World, Report 4, pp. 14–21, World Health Organization, Geneva
-
- Munro A. W., McLean K. J., Marshall K. R., Warman A. J., Lewis G., Roitel O., Sutcliffe M. J., Kemp C. A., Modi S., Scrutton N. S., Leys D. (2003) Biochem. Soc. Trans. 31, 625–630 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

