Insights into the reactivation of cobalamin-dependent methionine synthase
- PMID: 19846791
- PMCID: PMC2765455
- DOI: 10.1073/pnas.0906132106
Insights into the reactivation of cobalamin-dependent methionine synthase
Abstract
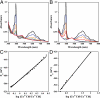
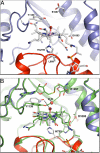
Cobalamin-dependent methionine synthase (MetH) is a modular protein that catalyzes the transfer of a methyl group from methyltetrahydrofolate to homocysteine to produce methionine and tetrahydrofolate. The cobalamin cofactor, which serves as both acceptor and donor of the methyl group, is oxidized once every approximately 2,000 catalytic cycles and must be reactivated by the uptake of an electron from reduced flavodoxin and a methyl group from S-adenosyl-L-methionine (AdoMet). Previous structures of a C-terminal fragment of MetH (MetH(CT)) revealed a reactivation conformation that juxtaposes the cobalamin- and AdoMet-binding domains. Here we describe 2 structures of a disulfide stabilized MetH(CT) ((s-s)MetH(CT)) that offer further insight into the reactivation of MetH. The structure of (s-s)MetH(CT) with cob(II)alamin and S-adenosyl-L-homocysteine represents the enzyme in the reactivation step preceding electron transfer from flavodoxin. The structure supports earlier suggestions that the enzyme acts to lower the reduction potential of the Co(II)/Co(I) couple by elongating the bond between the cobalt and its upper axial water ligand, effectively making the cobalt 4-coordinate, and illuminates the role of Tyr-1139 in the stabilization of this 4-coordinate state. The structure of (s-s)MetH(CT) with aquocobalamin may represent a transient state at the end of reactivation as the newly remethylated 5-coordinate methylcobalamin returns to the 6-coordinate state, triggering the rearrangement to a catalytic conformation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Goulding CW, Postigo D, Matthews RG. Cobalamin-dependent methionine synthase is a modular protein with distinct regions for binding homocysteine, methyltetrahydrofolate, cobalamin, and adenosylmethionine. Biochemistry. 1997;36:8082–8091. - PubMed
-
- Drummond JT, Huang S, Blumenthal RM, Matthews RG. Assignment of enzymatic function to specific protein regions of cobalamin-dependent methionine synthase from Escherichia coli. Biochemistry. 1993;32:9290–9295. - PubMed
-
- Fujii K, Galivan JH, Huennekens FM. Activation of methionine synthase: Further characterization of flavoprotein system. Arch Biochem Biophys. 1977;178:662–670. - PubMed
-
- Taylor RT, Weissbach H. N5-methyltetrahydrofolate-homocysteine transmethylase: Partial purification and properties. J Biol Chem. 1967;242:1502–1508. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

