Metabolomic profiling in LRRK2-related Parkinson's disease
- PMID: 19847307
- PMCID: PMC2761616
- DOI: 10.1371/journal.pone.0007551
Metabolomic profiling in LRRK2-related Parkinson's disease
Abstract
Background: Mutations in LRRK2 gene represent the most common known genetic cause of Parkinson's disease (PD).
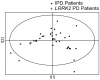
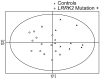
Methodology/principal findings: We used metabolomic profiling to identify biomarkers that are associated with idiopathic and LRRK2 PD. We compared plasma metabolomic profiles of patients with PD due to the G2019S LRRK2 mutation, to asymptomatic family members of these patients either with or without G2019S LRRK2 mutations, and to patients with idiopathic PD, as well as non-related control subjects. We found that metabolomic profiles of both idiopathic PD and LRRK2 PD subjects were clearly separated from controls. LRRK2 PD patients had metabolomic profiles distinguishable from those with idiopathic PD, and the profiles could predict whether the PD was secondary to LRRK2 mutations or idiopathic. Metabolomic profiles of LRRK2 PD patients were well separated from their family members, but there was a slight overlap between family members with and without LRRK2 mutations. Both LRRK2 and idiopathic PD patients showed significantly reduced uric acid levels. We also found a significant decrease in levels of hypoxanthine and in the ratios of major metabolites of the purine pathway in plasma of PD patients.
Conclusions/significance: These findings show that LRRK2 patients with the G2019S mutation have unique metabolomic profiles that distinguish them from patients with idiopathic PD. Furthermore, asymptomatic LRRK2 carriers can be separated from gene negative family members, which raises the possibility that metabolomic profiles could be useful in predicting which LRRK2 carriers will eventually develop PD. The results also suggest that there are aberrations in the purine pathway in PD which may occur upstream from uric acid.
Conflict of interest statement
Figures





References
-
- Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Genetics of Parkinson's disease and parkinsonism. Ann Neurol. 2006;60:389–398. - PubMed
-
- Klein C, Schlossmacher MG. Parkinson disease, 10 years after its genetic revolution: multiple clues to a complex disorder. Neurology. 2007;69:2093–2104. - PubMed
-
- Schiesling C, Kieper N, Seidel K, Kruger R. Review: Familial Parkinson's disease–genetics, clinical phenotype and neuropathology in relation to the common sporadic form of the disease. Neuropathol Appl Neurobiol. 2008;34:255–271. - PubMed
-
- Thomas B, Beal MF. Parkinson's disease. Hum Mol Genet. 2007;16 Spec No. 2:R183–194. - PubMed
-
- Bras JM, Guerreiro RJ, Ribeiro MH, Januario C, Morgadinho A, et al. G2019S dardarin substitution is a common cause of Parkinson's disease in a Portuguese cohort. Mov Disord. 2005;20:1653–1655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

