DDBJ launches a new archive database with analytical tools for next-generation sequence data
- PMID: 19850725
- PMCID: PMC2808917
- DOI: 10.1093/nar/gkp847
DDBJ launches a new archive database with analytical tools for next-generation sequence data
Abstract
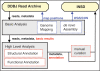
The DNA Data Bank of Japan (DDBJ) (http://www.ddbj.nig.ac.jp) has collected and released 1,701,110 entries/1,116,138,614 bases between July 2008 and June 2009. A few highlighted data releases from DDBJ were the complete genome sequence of an endosymbiont within protist cells in the termite gut and Cap Analysis Gene Expression tags for human and mouse deposited from the Functional Annotation of the Mammalian cDNA consortium. In this period, we started a novel user announcement service using Really Simple Syndication (RSS) to deliver a list of data released from DDBJ on a daily basis. Comprehensive visualization of a DDBJ release data was attempted by using a word cloud program. Moreover, a new archive for sequencing data from next-generation sequencers, the 'DDBJ Read Archive' (DRA), was launched. Concurrently, for read data registered in DRA, a semi-automatic annotation tool called the 'DDBJ Read Annotation Pipeline' was released as a preliminary step. The pipeline consists of two parts: basic analysis for reference genome mapping and de novo assembly and high-level analysis of structural and functional annotations. These new services will aid users' research and provide easier access to DDBJ databases.
Figures



References
-
- Hongoh Y, Sharma VK, Prakash T, Noda S, Toh H, Taylor TD, Kudo T, Sakaki Y, Toyoda A, Hattori M, et al. Genome of an endosymbiont coupling n2 fixation to cellulolysis within protist cells in termite gut. Science. 2008;322:1108–1109. - PubMed
-
- Cochrane G, Bates K, Apweiler R, Tateno Y, Mashima J, Kosuge T, Mizrachi IK, Schafer S, Fetchko M. Evidence standards in experimental and inferential INSDC third party annotation data. OMICS. 2006;10:105–113. - PubMed
-
- Kosuge T, Abe T, Okido T, Tanaka N, Hirahata M, Maruyama Y, Mashima J, Tomiki A, Kurokawa M, Himeno R, et al. Exploration and grading of possible genes in 183 bacterial strains by a common fine protocol lead to new genes: gene trek in prokaryote space (GTPS) DNA Res. 2006;13:245–254. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

