Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs
- PMID: 19850906
- PMCID: PMC2779667
- DOI: 10.1261/rna.1738409
Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs
Abstract
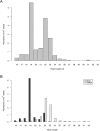
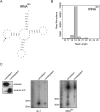
Deep sequencing technologies such as Illumina, SOLiD, and 454 platforms have become very powerful tools in discovering and quantifying small RNAs in diverse organisms. Sequencing small RNA fractions always identifies RNAs derived from abundant RNA species such as rRNAs, tRNAs, snRNA, and snoRNA, and they are widely considered to be random degradation products. We carried out bioinformatic analysis of deep sequenced HeLa RNA and after quality filtering, identified highly abundant small RNA fragments, derived from mature tRNAs that are likely produced by specific processing rather than from random degradation. Moreover, we showed that the processing of small RNAs derived from tRNA(Gln) is dependent on Dicer in vivo and that Dicer cleaves the tRNA in vitro.
Figures






References
-
- Abouelhoda MI, Kurtz S, Ohlebusch E. Replacing suffix trees with enhanced suffix trees. J Discrete Algorithms. 2004;2:53–86.
-
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
