Do conformational biases of simple helical junctions influence RNA folding stability and specificity?
- PMID: 19850914
- PMCID: PMC2779674
- DOI: 10.1261/rna.1747509
Do conformational biases of simple helical junctions influence RNA folding stability and specificity?
Abstract
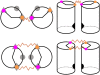
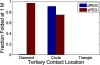
Structured RNAs must fold into their native structures and discriminate against a large number of alternative ones, an especially difficult task given the limited information content of RNA's nucleotide alphabet. The simplest motifs within structured RNAs are two helices joined by nonhelical junctions. To uncover the fundamental behavior of these motifs and to elucidate the underlying physical forces and challenges faced by structured RNAs, we computationally and experimentally studied a tethered duplex model system composed of two helices joined by flexible single- or double-stranded polyethylene glycol tethers, whose lengths correspond to those typically observed in junctions from structured RNAs. To dissect the thermodynamic properties of these simple motifs, we computationally probed how junction topology, electrostatics, and tertiary contact location influenced folding stability. Small-angle X-ray scattering was used to assess our predictions. Single- or double-stranded junctions, independent of sequence, greatly reduce the space of allowed helical conformations and influencing the preferred location and orientation of their adjoining helices. A double-stranded junction guides the helices along a hinge-like pathway. In contrast, a single-stranded junction samples a broader set of conformations and has different preferences than the double-stranded junction. In turn, these preferences determine the stability and distinct specificities of tertiary structure formation. These sequence-independent effects suggest that properties as simple as a junction's topology can generally define the accessible conformational space, thereby stabilizing desired structures and assisting in discriminating against misfolded structures. Thus, junction topology provides a fundamental strategy for transcending the limitations imposed by the low information content of RNA primary sequence.
Figures

 axis was chosen to point along the helical axis, while the
axis was chosen to point along the helical axis, while the  axis was chosen to point orthogonally relative to the
axis was chosen to point orthogonally relative to the  axis in the direction of the 3′ oxygen of the terminal residue. The
axis in the direction of the 3′ oxygen of the terminal residue. The  axis was perpendicular to the
axis was perpendicular to the  and
and  axes and computed as
axes and computed as  . As noted, the polar angle φ subtended by the 5′ and 3′ oxygens is approximately −133°.
. As noted, the polar angle φ subtended by the 5′ and 3′ oxygens is approximately −133°.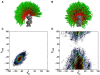


References
-
- Bowman AW, Azzalini A. Applied smoothing techniques for data analysis: The kernel approach with S-Plus illustrations. Oxford University Press; Oxford, UK: 1997.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
