Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers
- PMID: 19851283
- PMCID: PMC2797054
- DOI: 10.1038/emboj.2009.306
Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers
Abstract
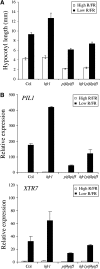
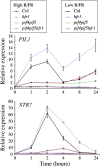
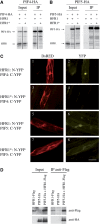
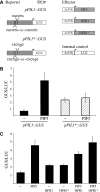
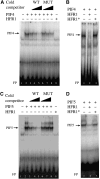
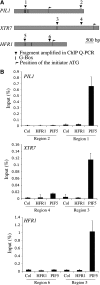
In shade-intolerant plants such as Arabidopsis, a reduction in the red/far-red (R/FR) ratio, indicative of competition from other plants, triggers a suite of responses known as the shade avoidance syndrome (SAS). The phytochrome photoreceptors measure the R/FR ratio and control the SAS. The phytochrome-interacting factors 4 and 5 (PIF4 and PIF5) are stabilized in the shade and are required for a full SAS, whereas the related bHLH factor HFR1 (long hypocotyl in FR light) is transcriptionally induced by shade and inhibits this response. Here we show that HFR1 interacts with PIF4 and PIF5 and limits their capacity to induce the expression of shade marker genes and to promote elongation growth. HFR1 directly inhibits these PIFs by forming non-DNA-binding heterodimers with PIF4 and PIF5. Our data indicate that PIF4 and PIF5 promote SAS by directly binding to G-boxes present in the promoter of shade marker genes, but their action is limited later in the shade when HFR1 accumulates and forms non-DNA-binding heterodimers. This negative feedback loop is important to limit the response of plants to shade.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Alabadi D, Blazquez MA (2009) Molecular interactions between light and hormone signaling to control plant growth. Plant Mol Biol 69: 409–417 - PubMed
-
- Alabadi D, Gallego-Bartolome J, Orlando L, Garcia-Carcel L, Rubio V, Martinez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, Blazquez MA (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 - PubMed
-
- Ballare CL (1999) Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci 4: 97–102 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

