Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK
- PMID: 19851285
- PMCID: PMC2790489
- DOI: 10.1038/emboj.2009.302
Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK
Abstract
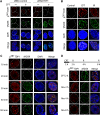
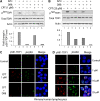
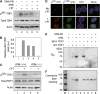
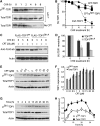
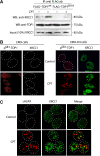
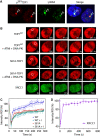
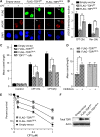
Human tyrosyl-DNA phosphodiesterase (TDP1) hydrolyzes the phosphodiester bond at a DNA 3' end linked to a tyrosyl moiety. This type of linkage is found at stalled topoisomerase I (Top1)-DNA covalent complexes, and TDP1 has been implicated in the repair of such complexes. Here we show that Top1-associated DNA double-stranded breaks (DSBs) induce the phosphorylation of TDP1 at S81. This phosphorylation is mediated by the protein kinases: ataxia-telangiectasia-mutated (ATM) and DNA-dependent protein kinase (DNA-PK). Phosphorylated TDP1 forms nuclear foci that co-localize with those of phosphorylated histone H2AX (gammaH2AX). Both Top1-induced replication- and transcription-mediated DNA damages induce TDP1 phosphorylation. Furthermore, we show that S81 phosphorylation stabilizes TDP1, induces the formation of XRCC1 (X-ray cross-complementing group 1)-TDP1 complexes and enhances the mobilization of TDP1 to DNA damage sites. Finally, we provide evidence that TDP1-S81 phosphorylation promotes cell survival and DNA repair in response to CPT-induced DSBs. Together; our findings provide a new mechanism for TDP1 post-translational regulation by ATM and DNA-PK.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Abraham RT (2004) PI 3-kinase-related kinases: ‘big' players in stress-induced signaling pathways. DNA Repair (Amst) 3: 883–887 - PubMed
-
- Antony S, Marchand C, Stephen AG, Thibaut L, Agama KK, Fisher RJ, Pommier Y (2007) Novel high-throughput electrochemiluminescent assay for identification of human tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1. Nucleic Acids Res 35: 4474–4484 - PMC - PubMed
-
- Audebert M, Salles B, Calsou P (2004) Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 279: 55117–55126 - PubMed
-
- Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

