Self-regulation of Stat3 activity coordinates cell-cycle progression and neural crest specification
- PMID: 19851287
- PMCID: PMC2808363
- DOI: 10.1038/emboj.2009.313
Self-regulation of Stat3 activity coordinates cell-cycle progression and neural crest specification
Abstract
A complex set of extracellular signals is required for neural crest (NC) specification. However, how these signals function to coordinate cell-cycle progression and differentiation remains poorly understood. Here, we report in Xenopus a role for the transcription factor signal transducers and activators of transcription-3 (Stat3) in this process downstream of FGF signalling. Depletion of Stat3 inhibits NC gene expression and cell proliferation, whereas overexpression expands the NC domain and promotes cell proliferation. Stat3 is phosphorylated and activated in ectodermal cells by FGFs through binding with FGFR4. Stat3 activation is also modulated by Hairy2 and Id3 proteins that, respectively, facilitate and disrupt Stat3-FGFR4 complex formation. Furthermore, distinct levels of Stat3 activity control Hairy2 and Id3 transcription, leading to Stat3 self-regulation. Finally, high Stat3 activity maintains cells in an undifferentiated state, whereas low activity promotes cell proliferation and NC differentiation. Together, our data suggest that Stat3, downstream of FGFs and under the positive and negative feedback regulation of Hairy2 and Id3, plays an essential role in the coordination of cell-cycle progression and differentiation during NC specification.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

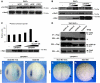
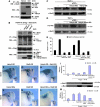




References
-
- Ahrens K, Schlosser G (2005) Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev Biol 288: 40–59 - PubMed
-
- Amaya E, Musci TJ, Kirschner MW (1991) Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell 66: 257–270 - PubMed
-
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP (2003) Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev Biol 259: 19–33 - PubMed
-
- Bellmeyer A, Krase J, Lindgren J, LaBonne C (2003) The protooncogene c-myc is an essential regulator of neural crest formation in Xenopus. Dev Cell 4: 827–839 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

