Building ubiquitin chains: E2 enzymes at work
- PMID: 19851334
- PMCID: PMC3107738
- DOI: 10.1038/nrm2780
Building ubiquitin chains: E2 enzymes at work
Abstract
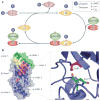
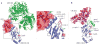
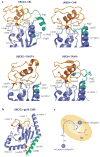
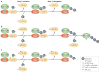
The modification of proteins with ubiquitin chains can change their localization, activity and/or stability. Although ubiquitylation requires the concerted action of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s), it is the E2s that have recently emerged as key mediators of chain assembly. These enzymes are able to govern the switch from ubiquitin chain initiation to elongation, regulate the processivity of chain formation and establish the topology of assembled chains, thereby determining the consequences of ubiquitylation for the modified proteins.
Figures

References
-
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. - PubMed
-
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. - PubMed
-
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. - PubMed
-
- Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nature Cell Biol. 2009;11:123–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

