An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations
- PMID: 19851441
- PMCID: PMC2757866
- DOI: 10.1371/journal.pcbi.1000541
An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations
Abstract
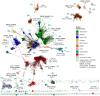
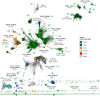
The group of proteins that contain a thioredoxin (Trx) fold is huge and diverse. Assessment of the variation in catalytic machinery of Trx fold proteins is essential in providing a foundation for understanding their functional diversity and predicting the function of the many uncharacterized members of the class. The proteins of the Trx fold class retain common features-including variations on a dithiol CxxC active site motif-that lead to delivery of function. We use protein similarity networks to guide an analysis of how structural and sequence motifs track with catalytic function and taxonomic categories for 4,082 representative sequences spanning the known superfamilies of the Trx fold. Domain structure in the fold class is varied and modular, with 2.8% of sequences containing more than one Trx fold domain. Most member proteins are bacterial. The fold class exhibits many modifications to the CxxC active site motif-only 56.8% of proteins have both cysteines, and no functional groupings have absolute conservation of the expected catalytic motif. Only a small fraction of Trx fold sequences have been functionally characterized. This work provides a global view of the complex distribution of domains and catalytic machinery throughout the fold class, showing that each superfamily contains remnants of the CxxC active site. The unifying context provided by this work can guide the comparison of members of different Trx fold superfamilies to gain insight about their structure-function relationships, illustrated here with the thioredoxins and peroxiredoxins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. - PubMed
-
- Bashton M, Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. - PubMed
-
- Krishna SS, Grishin NV. Structural drift: a possible path to protein fold change. Bioinformatics. 2005;21:1308–1310. - PubMed
-
- Qi Y, Grishin NV. Structural classification of thioredoxin-like fold proteins. Proteins. 2005;58:376–388. - PubMed
-
- Martin JL. Thioredoxin–a fold for all reasons. Structure. 1995;3:245–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

