Quality control in microarray assessment of gene expression in human airway epithelium
- PMID: 19852842
- PMCID: PMC2774870
- DOI: 10.1186/1471-2164-10-493
Quality control in microarray assessment of gene expression in human airway epithelium
Abstract
Background: Microarray technology provides a powerful tool for defining gene expression profiles of airway epithelium that lend insight into the pathogenesis of human airway disorders. The focus of this study was to establish rigorous quality control parameters to ensure that microarray assessment of the airway epithelium is not confounded by experimental artifact. Samples (total n = 223) of trachea, large and small airway epithelium were collected by fiberoptic bronchoscopy of 144 individuals and hybridized to Affymetrix microarrays. The pre- and post-chip quality control (QC) criteria established, included: (1) RNA quality, assessed by RNA Integrity Number (RIN) > or = 7.0; (2) cRNA transcript integrity, assessed by signal intensity ratio of GAPDH 3' to 5' probe sets < or = 3.0; and (3) the multi-chip normalization scaling factor < or = 10.0.
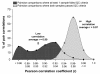
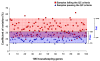
Results: Of the 223 samples, all three criteria were assessed in 191; of these 184 (96.3%) passed all three criteria. For the remaining 32 samples, the RIN was not available, and only the other two criteria were used; of these 29 (90.6%) passed these two criteria. Correlation coefficients for pairwise comparisons of expression levels for 100 maintenance genes in which at least one array failed the QC criteria (average Pearson r = 0.90 +/- 0.04) were significantly lower (p < 0.0001) than correlation coefficients for pairwise comparisons between arrays that passed the QC criteria (average Pearson r = 0.97 +/- 0.01). Inter-array variability was significantly decreased (p < 0.0001) among samples passing the QC criteria compared with samples failing the QC criteria.
Conclusion: Based on the aberrant maintenance gene data generated from samples failing the established QC criteria, we propose that the QC criteria outlined in this study can accurately distinguish high quality from low quality data, and can be used to delete poor quality microarray samples before proceeding to higher-order biological analyses and interpretation.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

