The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB
- PMID: 19853611
- PMCID: PMC4335309
- DOI: 10.1016/j.jmb.2009.10.040
The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB
Abstract
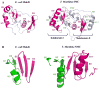
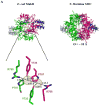
MukB, a divergent structural maintenance of chromosomes (SMC) protein, is important for chromosomal segregation and condensation in gamma-proteobacteria. MukB and canonical SMC proteins share a characteristic five-domain structure. Globular N- and C-terminal domains interact to form an ATP-binding cassette-like ATPase or "head" domain, which is connected to a smaller dimerization or "hinge" domain by a long, antiparallel coiled coil. In addition to mediating dimerization, this hinge region has been implicated in both conformational flexibility and dynamic protein-DNA interactions. We report here the first crystallographic model of the MukB hinge domain. This model also contains approximately 20% of the coiled-coil domain, including an unusual coiled-coil deviation. These results will facilitate studies to clarify the roles of both the hinge and the coiled-coil domains in MukB function.
Figures



Similar articles
-
A repeated coiled-coil interruption in the Escherichia coli condensin MukB.J Mol Biol. 2011 Dec 9;414(4):578-95. doi: 10.1016/j.jmb.2011.10.028. Epub 2011 Oct 25. J Mol Biol. 2011. PMID: 22041452
-
Identification of interacting regions within the coiled coil of the Escherichia coli structural maintenance of chromosomes protein MukB.J Mol Biol. 2009 Aug 7;391(1):57-73. doi: 10.1016/j.jmb.2009.05.070. Epub 2009 May 29. J Mol Biol. 2009. PMID: 19482037
-
Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications.Proteins. 2010 May 1;78(6):1483-90. doi: 10.1002/prot.22664. Proteins. 2010. PMID: 20034111
-
The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes.FEMS Microbiol Rev. 2014 May;38(3):380-92. doi: 10.1111/1574-6976.12045. Epub 2013 Nov 18. FEMS Microbiol Rev. 2014. PMID: 24118085 Free PMC article. Review.
-
A new twist in the coil: functions of the coiled-coil domain of structural maintenance of chromosome (SMC) proteins.Curr Genet. 2018 Feb;64(1):109-116. doi: 10.1007/s00294-017-0735-2. Epub 2017 Aug 23. Curr Genet. 2018. PMID: 28835994 Review.
Cited by
-
The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV.J Biol Chem. 2013 Mar 15;288(11):7653-7661. doi: 10.1074/jbc.M112.418087. Epub 2013 Jan 24. J Biol Chem. 2013. PMID: 23349462 Free PMC article.
-
Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.Mol Cell. 2021 Dec 2;81(23):4891-4906.e8. doi: 10.1016/j.molcel.2021.10.011. Epub 2021 Nov 4. Mol Cell. 2021. PMID: 34739874 Free PMC article.
-
Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo.EMBO J. 2013 Nov 13;32(22):2950-62. doi: 10.1038/emboj.2013.218. Epub 2013 Oct 4. EMBO J. 2013. PMID: 24097060 Free PMC article.
-
A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion.EMBO J. 2011 Jan 19;30(2):364-78. doi: 10.1038/emboj.2010.315. Epub 2010 Dec 7. EMBO J. 2011. PMID: 21139566 Free PMC article.
-
The bacterial condensin MukB compacts DNA by sequestering supercoils and stabilizing topologically isolated loops.J Biol Chem. 2017 Oct 13;292(41):16904-16920. doi: 10.1074/jbc.M117.803312. Epub 2017 Aug 25. J Biol Chem. 2017. PMID: 28842486 Free PMC article.
References
-
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. - PubMed
-
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. - PubMed
-
- Cobbe N, Heck MMS. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21:332–347. - PubMed
-
- Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. - PubMed
-
- Soppa J. Prokaryotic structural maintenance of chromosomes (SMC) proteins: distribution, phylogeny, and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene. 2001;278:253–264. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

