Exploring the linkage dependence of polyubiquitin conformations using molecular modeling
- PMID: 19853612
- PMCID: PMC2813430
- DOI: 10.1016/j.jmb.2009.10.039
Exploring the linkage dependence of polyubiquitin conformations using molecular modeling
Abstract
Posttranslational modification of proteins by covalent attachment of a small protein ubiquitin (Ub) or a polymeric chain of Ub molecules (called polyubiquitin) is involved in controlling a vast variety of processes in eukaryotic cells. The question of how different polyubiquitin signals are recognized is central to understanding the specificity of various types of polyubiquitination. In polyubiquitin, monomers are linked to each other via an isopeptide bond between the C-terminal glycine of one Ub and a lysine of the other. The functional outcome of polyubiquitination depends on the particular lysine involved in chain formation and appears to rely on linkage-dependent conformation of polyubiquitin. Thus, K48-linked chains, a universal signal for proteasomal degradation, under physiological conditions adopt a closed conformation where functionally important residues L8, I44, and V70 are sequestered at the interface between two adjacent Ub monomers. By contrast, K63-linked chains, which act as a nonproteolytic regulatory signal, adopt an extended conformation that lacks hydrophobic interubiquitin contact. Little is known about the functional roles of the so-called "noncanonical" chains (linked via K6, K11, K27, K29, or K33, or linked head-to-tail), and no structural information on these chains is available, except for information on the crystal structure of the head-to-tail-linked diubiquitin (Ub(2)). In this study, we use molecular modeling to examine whether any of the noncanonical chains can adopt a closed conformation similar to that in K48-linked polyubiquitin. Our results show that the eight possible Ub(2) chains can be divided into two groups: chains linked via K6, K11, K27, or K48 are predicted to form a closed conformation, whereas chains linked via K29, K33, or K63, or linked head-to-tail are unable to form such a contact due to steric occlusion. These predictions are validated by the known structures of K48-, K63-, and head-to-tail-linked chains. Our study also predicts structural models for Ub(2) chains linked via K6, K11, or K27. The implications of these findings for linkage-selective recognition of noncanonical polyubiquitin signals by various receptors are discussed.
Figures
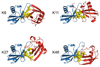
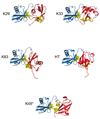

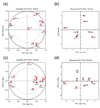
References
-
- Wilkinson K. Ubiquitin: a Nobel protein. Cell. 2004;119:741–745. - PubMed
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. - PubMed
-
- Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–16471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

