Biased chromatin signatures around polyadenylation sites and exons
- PMID: 19854133
- PMCID: PMC2786773
- DOI: 10.1016/j.molcel.2009.10.008
Biased chromatin signatures around polyadenylation sites and exons
Abstract
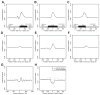
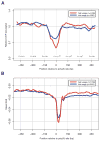
Core RNA-processing reactions in eukaryotic cells occur cotranscriptionally in a chromatin context, but the relationship between chromatin structure and pre-mRNA processing is poorly understood. We observed strong nucleosome depletion around human polyadenylation sites (PAS) and nucleosome enrichment just downstream of PAS. In genes with multiple alternative PAS, higher downstream nucleosome affinity was associated with higher PAS usage, independently of known PAS motifs that function at the RNA level. Conversely, exons were associated with distinct peaks in nucleosome density. Exons flanked by long introns or weak splice sites exhibited stronger nucleosome enrichment, and incorporation of nucleosome density data improved splicing simulation accuracy. Certain histone modifications, including H3K36me3 and H3K27me2, were specifically enriched on exons, suggesting active marking of exon locations at the chromatin level. Together, these findings provide evidence for extensive functional connections between chromatin structure and RNA processing.
Figures


References
-
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. - PubMed
-
- Baldi P, Brunak S, Chauvin Y, Krogh A. Naturally occurring nucleosome positioning signals in human exons and introns. J Mol Biol. 1996;263:503–510. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

