Mutations at the same amino acid in myosin that cause either skeletal or cardiac myopathy have distinct molecular phenotypes
- PMID: 19854198
- PMCID: PMC2854248
- DOI: 10.1016/j.yjmcc.2009.10.011
Mutations at the same amino acid in myosin that cause either skeletal or cardiac myopathy have distinct molecular phenotypes
Abstract
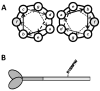
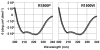
To date, more than 230 disease-causing mutations have been linked to the slow/cardiac muscle myosin gene, beta-MyHC (MYH7). Most of these mutations are located in the globular head region of the protein and result in cardiomyopathies. Recently, however, a number of novel disease-causing mutations have been described in the long, alpha-helical, coiled coil tail region of the beta-MyHC protein. Mutations in this region are of particular interest because they are associated with a multitude of human diseases, including both cardiac and skeletal myopathies. Here, we attempt to dissect the mechanism(s) by which mutations in the rod region of beta-MyHC can cause a variety of diseases by analyzing two mutations at a single amino acid (R1500P and R1500W) which cause two distinct diseases (Laing-type early-onset distal myopathy and dilated cardiomyopathy, respectively). For diseases linked to the R1500 residue, we find that each mutation displays distinct structural, thermodynamic, and functional properties. Both R1500P and R1500W cause a decrease in thermodynamic stability, although the R1500W phenotype is more severe. Both mutations also affect filament assembly, with R1500P causing an additional decrease in filament stability. In addition to furthering our understanding of the mechanism of pathogenesis for each of these diseases, these data also suggest how the variance in molecular phenotype may be associated with the variance in clinical phenotype present with mutations in the beta-MyHC rod.
Copyright (c) 2009 Elsevier Ltd. All rights reserved.
Figures

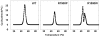


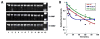
References
-
- McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982 Sep 16;299(5880):226–31. - PubMed
-
- Atkinson SJ, Stewart M. Molecular interactions in myosin assembly. Role of the 28-residue charge repeat in the rod. J Mol Biol. 1992 Jul 5;226(1):7–13. - PubMed
-
- Atkinson SJ, Stewart M. Molecular basis of myosin assembly: coiled-coil interactions and the role of charge periodicities. J Cell Sci Suppl. 1991;14:7–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

