Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus
- PMID: 19854896
- PMCID: PMC2798248
- DOI: 10.1128/JB.01202-09
Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus
Abstract
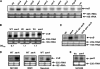
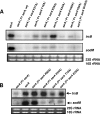
Thioredoxin reductase (encoded by trxB) protects Staphylococcus aureus against oxygen or disulfide stress and is indispensable for growth. Among the different sarA family mutants analyzed, transcription of trxB was markedly elevated in the sarA mutant under conditions of aerobic as well as microaerophilic growth, indicating that SarA acts as a negative regulator of trxB expression. Gel shift analysis showed that purified SarA protein binds directly to the trxB promoter region DNA in vitro. DNA binding of SarA was essential for repression of trxB transcription in vivo in S. aureus. Northern blot analysis and DNA binding studies of the purified wild-type SarA and the mutant SarAC9G with oxidizing agents indicated that oxidation of Cys-9 reduced the binding of SarA to the trxB promoter DNA. Oxidizing agents, in particular diamide, could further enhance transcription of the trxB gene in the sarA mutant, suggesting the presence of a SarA-independent mode of trxB induction. Analysis of two oxidative stress-responsive sarA regulatory target genes, trxB and sodM, with various mutant sarA constructs showed a differential ability of the SarA to regulate expression of the two above-mentioned genes in vivo. The overall data demonstrate the important role played by SarA in modulating expression of genes involved in oxidative stress resistance in S. aureus.
Figures



References
-
- Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. - PubMed
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagal, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

