Combined genomic and proteomic approaches identify gene clusters involved in anaerobic 2-methylnaphthalene degradation in the sulfate-reducing enrichment culture N47
- PMID: 19854898
- PMCID: PMC2798259
- DOI: 10.1128/JB.00874-09
Combined genomic and proteomic approaches identify gene clusters involved in anaerobic 2-methylnaphthalene degradation in the sulfate-reducing enrichment culture N47
Abstract
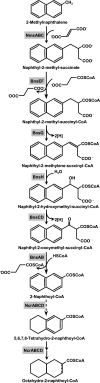
The highly enriched deltaproteobacterial culture N47 anaerobically oxidizes the polycyclic aromatic hydrocarbons naphthalene and 2-methylnaphthalene, with sulfate as the electron acceptor. Combined genome sequencing and liquid chromatography-tandem mass spectrometry-based shotgun proteome analyses were performed to identify genes and proteins involved in anaerobic aromatic catabolism. Proteome analysis of 2-methylnaphthalene-grown N47 cells resulted in the identification of putative enzymes catalyzing the anaerobic conversion of 2-methylnaphthalene to 2-naphthoyl coenzyme A (2-naphthoyl-CoA), as well as the reductive ring cleavage of 2-naphthoyl-CoA, leading to the formation of acetyl-CoA and CO(2). The glycyl radical-catalyzed fumarate addition to the methyl group of 2-methylnaphthalene is catalyzed by naphthyl-2-methyl-succinate synthase (Nms), composed of alpha-, beta-, and gamma-subunits that are encoded by the genes nmsABC. Located upstream of nmsABC is nmsD, encoding the Nms-activating enzyme, which harbors the characteristic [Fe(4)S(4)] cluster sequence motifs of S-adenosylmethionine radical enzymes. The bns gene cluster, coding for enzymes involved in beta-oxidation reactions converting naphthyl-2-methyl-succinate to 2-naphthoyl-CoA, was found four intervening open reading frames further downstream. This cluster consists of eight genes (bnsABCDEFGH) corresponding to 8.1 kb, which are closely related to genes for enzymes involved in anaerobic toluene degradation within the denitrifiers "Aromatoleum aromaticum" EbN1, Azoarcus sp. strain T, and Thauera aromatica. Another contiguous DNA sequence harbors the gene for 2-naphthoyl-CoA reductase (ncr) and 16 additional genes that were found to be expressed in 2-methylnaphthalene-grown cells. These genes code for enzymes that were supposed to catalyze the dearomatization and ring cleavage reactions converting 2-naphthoyl-CoA to acetyl-CoA and CO(2). Comparative sequence analysis of the four encoding subunits (ncrABCD) showed the gene product to have the closest similarity to the Azoarcus type of benzoyl-CoA reductase. The present work provides the first insight into the genetic basis of anaerobic 2-methylnaphthalene metabolism and delivers implications for understanding contaminant degradation.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

