Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon
- PMID: 19854902
- PMCID: PMC2798271
- DOI: 10.1128/JB.00741-09
Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon
Abstract
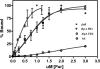
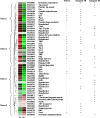
To ensure survival in the host, bacteria have evolved strategies to acquire the essential element iron. In Neisseria gonorrhoeae, the ferric uptake regulator Fur regulates metabolism through transcriptional control of iron-responsive genes by binding conserved Fur box (FB) sequences in promoters during iron-replete growth. Our previous studies showed that Fur also controls the transcription of secondary regulators that may, in turn, control pathways important to pathogenesis, indicating an indirect role for Fur in controlling these downstream genes. To better define the iron-regulated cascade of transcriptional control, we combined three global strategies--temporal transcriptome analysis, genomewide in silico FB prediction, and Fur titration assays (FURTA)--to detect genomic regions able to bind Fur in vivo. The majority of the 300 iron-repressed genes were predicted to be of unknown function, followed by genes involved in iron metabolism, cell communication, and intermediary metabolism. The 107 iron-induced genes encoded hypothetical proteins or energy metabolism functions. We found 28 predicted FBs in FURTA-positive clones in the promoters and within the open reading frames of iron-repressed genes. We found lower levels of conservation at critical thymidine residues involved in Fur binding in the FB sequence logos of FURTA-positive clones with intragenic FBs than in the sequence logos generated from FURTA-positive promoter regions. In electrophoretic mobility shift assay studies, intragenic FBs bound Fur with a lower affinity than intergenic FBs. Our findings further indicate that transcription under iron stress is indirectly controlled by Fur through 12 potential secondary regulators.
Figures




References
-
- Andrews, S. C., A. K. Robinson, and F. Rodríguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. - PubMed
-
- Angerer, A., and V. Braun. 1998. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 169:483-490. - PubMed
-
- Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

