Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters
- PMID: 19854911
- PMCID: PMC2798258
- DOI: 10.1128/JB.01124-09
Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters
Abstract
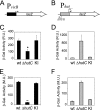
Type IV secretion systems (T4SS) are multicomponent machineries involved in the translocation of effector molecules across the bacterial cell envelope. The virB operon of Brucella abortus codes for a T4SS that is essential for virulence and intracellular multiplication of the bacterium in the host. Previous studies showed that the virB operon of B. abortus is tightly regulated within the host cells. In order to identify factors implicated in the control of virB expression, we searched for proteins of Brucella that directly bind to the virB promoter (P(virB)). Using different procedures, we isolated a 27-kDa protein that binds specifically to P(virB). This protein was identified as HutC, the transcriptional repressor of the histidine utilization (hut) genes. Analyses of virB and hut promoter activity revealed that HutC exerts two different roles: it acts as a coactivator of transcription of the virB operon, whereas it represses the hut genes. Such activities were observed both intracellularly and in bacteria incubated under conditions that resemble the intracellular environment. Electrophoresis mobility shift assays (EMSA) and DNase I footprinting experiments revealed the structure, affinity, and localization of the HutC-binding sites and supported the regulatory role of HutC in both hut and virB promoters. Taken together, these results indicate that Brucella coopted the function of HutC to coordinate the Hut pathway with transcriptional regulation of the virB genes, probably as a way to sense its own metabolic state and develop adaptive responses to overcome intracellular host defenses.
Figures
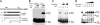
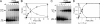

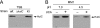
References
-
- Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U. S. A. 99:1544-1549. - PMC - PubMed
-
- Broughton, E. S., and K. L. Jahans. 1997. The differentiation of Brucella species by substrate specific tetrazolium reduction. Vet. Microbiol. 57:253-271. - PubMed
-
- Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. - PubMed
-
- Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159-168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

