Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner
- PMID: 19858288
- PMCID: PMC2798288
- DOI: 10.1128/MCB.00651-09
Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner
Abstract
mRNAs encoding mitochondrial proteins are enriched in the vicinity of mitochondria, presumably to facilitate protein transport. A possible mechanism for enrichment may involve interaction of the translocase of the mitochondrial outer membrane (TOM) complex with the precursor protein while it is translated, thereby leading to association of polysomal mRNAs with mitochondria. To test this hypothesis, we isolated mitochondrial fractions from yeast cells lacking the major import receptor, Tom20, and compared their mRNA repertoire to that of wild-type cells by DNA microarrays. Most mRNAs encoding mitochondrial proteins were less associated with mitochondria, yet the extent of decrease varied among genes. Analysis of several mRNAs revealed that optimal association of Tom20 target mRNAs requires both translating ribosomes and features within the encoded mitochondrial targeting signal. Recently, Puf3p was implicated in the association of mRNAs with mitochondria through interaction with untranslated regions. We therefore constructed a tom20 Delta puf3 Delta double-knockout strain, which demonstrated growth defects under conditions where fully functional mitochondria are required. Mislocalization effects for few tested mRNAs appeared stronger in the double knockout than in the tom20 Delta strain. Taken together, our data reveal a large-scale mRNA association mode that involves interaction of Tom20p with the translated mitochondrial targeting sequence and may be assisted by Puf3p.
Figures


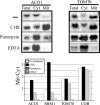
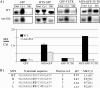

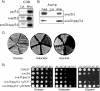

References
-
- Brandina, I., J. Graham, C. Lemaitre-Guillier, N. Entelis, I. Krasheninnikov, L. Sweetlove, I. Tarassov, and R. P. Martin. 2006. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim. Biophys. Acta 1757:1217-1228. - PubMed
-
- Czaplinski, K., and R. H. Singer. 2006. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 31:687-693. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
