Identity of the elusive IgM Fc receptor (FcmuR) in humans
- PMID: 19858324
- PMCID: PMC2806608
- DOI: 10.1084/jem.20091107
Identity of the elusive IgM Fc receptor (FcmuR) in humans
Abstract
Although Fc receptors (FcRs) for switched immunoglobulin (Ig) isotypes have been extensively characterized, FcR for IgM (FcmuR) has defied identification. By retroviral expression and functional cloning, we have identified a complementary DNA (cDNA) encoding a bona fide FcmuR in human B-lineage cDNA libraries. FcmuR is defined as a transmembrane sialoglycoprotein of approximately 60 kD, which contains an extracellular Ig-like domain homologous to two other IgM-binding receptors (polymeric Ig receptor and Fcalpha/muR) but exhibits an exclusive Fcmu-binding specificity. The cytoplasmic tail of FcmuR contains conserved Ser and Tyr residues, but none of the Tyr residues match the immunoreceptor tyrosine-based activation, inhibitory, or switch motifs. Unlike other FcRs, the major cell types expressing FcmuR are adaptive immune cells, including B and T lymphocytes. After antigen-receptor ligation or phorbol myristate acetate stimulation, FcmuR expression was up-regulated on B cells but was down-modulated on T cells, suggesting differential regulation of FcmuR expression during B and T cell activation. Although this receptor was initially designated as Fas apoptotic inhibitory molecule 3, or TOSO, our results indicate that FcmuR per se has no inhibitory activity in Fas-mediated apoptosis and that such inhibition is only achieved when anti-Fas antibody of an IgM but not IgG isotype is used for inducing apoptosis.
Figures
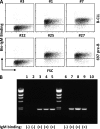
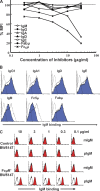




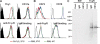

References
-
- Arase H., Saito T., Phillips J.H., Lanier L.L. 2001. Cutting edge: the mouse NK cell-associated antigen recognized by DX5 monoclonal antibody is CD49b (α 2 integrin, very late antigen-2). J. Immunol. 167:1141–1144 - PubMed
-
- Boes M., Esau C., Fischer M.B., Schmidt T., Carroll M., Chen J. 1998a. Enhanced B-1 cell development, but impaired IgG antibody responses in mice deficient in secreted IgM. J. Immunol. 160:4776–4787 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

