LACTB is a filament-forming protein localized in mitochondria
- PMID: 19858488
- PMCID: PMC2767363
- DOI: 10.1073/pnas.0906734106
LACTB is a filament-forming protein localized in mitochondria
Abstract
LACTB is a mammalian active-site serine protein that has evolved from a bacterial penicillin-binding protein. Penicillin-binding proteins are involved in the metabolism of peptidoglycan, the major bacterial cell wall constituent, implying that LACTB has been endowed with novel biochemical properties during eukaryote evolution. Here we demonstrate that LACTB is localized in the mitochondrial intermembrane space, where it is polymerized into stable filaments with a length extending more than a hundred nanometers. We infer that LACTB, through polymerization, promotes intramitochondrial membrane organization and micro-compartmentalization. These findings have implications for our understanding of mitochondrial evolution and function.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
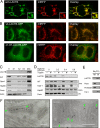

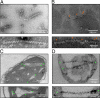

References
-
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. - PubMed
-
- de Duve C. The origin of eukaryotes: A reappraisal. Nat Rev Genet. 2007;8:395–403. - PubMed
-
- Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: Key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev. 2006;30:673–691. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

