Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila
- PMID: 19859525
- PMCID: PMC2760206
- DOI: 10.1371/journal.pbio.1000234
Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila
Abstract
Postzygotic reproductive barriers such as sterility and lethality of hybrids are important for establishing and maintaining reproductive isolation between species. Identifying the causal loci and discerning how they interfere with the development of hybrids is essential for understanding how hybrid incompatibilities (HIs) evolve, but little is known about the mechanisms of how HI genes cause hybrid dysfunctions. A previously discovered Drosophila melanogaster locus called Zhr causes lethality in F1 daughters from crosses between Drosophila simulans females and D. melanogaster males. Zhr maps to a heterochromatic region of the D. melanogaster X that contains 359-bp satellite repeats, suggesting either that Zhr is a rare protein-coding gene embedded within heterochromatin, or is a locus consisting of the noncoding repetitive DNA that forms heterochromatin. The latter possibility raises the question of how heterochromatic DNA can induce lethality in hybrids. Here we show that hybrid females die because of widespread mitotic defects induced by lagging chromatin at the time during early embryogenesis when heterochromatin is first established. The lagging chromatin is confined solely to the paternally inherited D. melanogaster X chromatids, and consists predominantly of DNA from the 359-bp satellite block. We further found that a rearranged X chromosome carrying a deletion of the entire 359-bp satellite block segregated normally, while a translocation of the 359-bp satellite block to the Y chromosome resulted in defective Y segregation in males, strongly suggesting that the 359-bp satellite block specifically and directly inhibits chromatid separation. In hybrids produced from wild-type parents, the 359-bp satellite block was highly stretched and abnormally enriched with Topoisomerase II throughout mitosis. The 359-bp satellite block is not present in D. simulans, suggesting that lethality is caused by the absence or divergence of factors in the D. simulans maternal cytoplasm that are required for heterochromatin formation of this species-specific satellite block. These findings demonstrate how divergence of noncoding repetitive sequences between species can directly cause reproductive isolation by altering chromosome segregation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


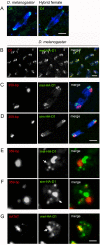

Comment in
-
Heterochromatin: a rapidly evolving species barrier.PLoS Biol. 2009 Oct;7(10):e1000233. doi: 10.1371/journal.pbio.1000233. Epub 2009 Sep 27. PLoS Biol. 2009. PMID: 19830257 Free PMC article.
References
-
- Ting C-T, Tsaur S-C, Wu M-L, Wu C-I. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. - PubMed
-
- Presgraves D. C, Balagopalan L, Abmayr S. M, Orr H. A. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423:715–719. - PubMed
-
- Barbash D. A, Awadalla P, Tarone A. M. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2004;2:e142. doi: 10.1371/journal.pbio.0020142. - DOI - PMC - PubMed
-
- Lee H-Y, Chou J-Y, Cheong L, Chang N-H, Yang S-Y, et al. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–1073. - PubMed
-
- Masly J. P, Jones C. D, Noor M. A, Locke J, Orr H. A. Gene transposition as a cause of hybrid sterility in Drosophila. Science. 2006;313:1448–1450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

