Structural genomics target selection for the New York consortium on membrane protein structure
- PMID: 19859826
- PMCID: PMC2780672
- DOI: 10.1007/s10969-009-9071-1
Structural genomics target selection for the New York consortium on membrane protein structure
Abstract
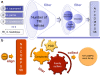
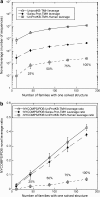
The New York Consortium on Membrane Protein Structure (NYCOMPS), a part of the Protein Structure Initiative (PSI) in the USA, has as its mission to establish a high-throughput pipeline for determination of novel integral membrane protein structures. Here we describe our current target selection protocol, which applies structural genomics approaches informed by the collective experience of our team of investigators. We first extract all annotated proteins from our reagent genomes, i.e. the 96 fully sequenced prokaryotic genomes from which we clone DNA. We filter this initial pool of sequences and obtain a list of valid targets. NYCOMPS defines valid targets as those that, among other features, have at least two predicted transmembrane helices, no predicted long disordered regions and, except for community nominated targets, no significant sequence similarity in the predicted transmembrane region to any known protein structure. Proteins that feed our experimental pipeline are selected by defining a protein seed and searching the set of all valid targets for proteins that are likely to have a transmembrane region structurally similar to that of the seed. We require sequence similarity aligning at least half of the predicted transmembrane region of seed and target. Seeds are selected according to their feasibility and/or biological interest, and they include both centrally selected targets and community nominated targets. As of December 2008, over 6,000 targets have been selected and are currently being processed by the experimental pipeline. We discuss how our target list may impact structural coverage of the membrane protein space.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

