Developing a set of ancestry-sensitive DNA markers reflecting continental origins of humans
- PMID: 19860882
- PMCID: PMC2775748
- DOI: 10.1186/1471-2156-10-69
Developing a set of ancestry-sensitive DNA markers reflecting continental origins of humans
Abstract
Background: The identification and use of Ancestry-Sensitive Markers (ASMs), i.e. genetic polymorphisms facilitating the genetic reconstruction of geographical origins of individuals, is far from straightforward.
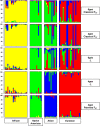
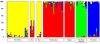
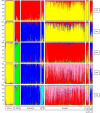
Results: Here we describe the ascertainment and application of five different sets of 47 single nucleotide polymorphisms (SNPs) allowing the inference of major human groups of different continental origin. For this, we first used 74 cell lines, representing human males from six different geographical areas and screened them with the Affymetrix Mapping 10K assay. In addition to using summary statistics estimating the genetic diversity among multiple groups of individuals defined by geography or language, we also used the program STRUCTURE to detect genetically distinct subgroups. Subsequently, we used a pairwise F(ST) ranking procedure among all pairs of genetic subgroups in order to identify a single best performing set of ASMs. Our initial results were independently confirmed by genotyping this set of ASMs in 22 individuals from Somalia, Afghanistan and Sudan and in 919 samples from the CEPH Human Genome Diversity Panel (HGDP-CEPH) CONCLUSION: By means of our pairwise population F(ST) ranking approach we identified a set of 47 SNPs that could serve as a panel of ASMs at a continental level.
Figures


References
-
- Frudakis T, Venkateswarlu K, Thomas MJ, Gaskin Z, Ginjupalli S, Gunturi S, Ponnuswamy V, Natarajan S, Nachimuthu PK. A classifier for the SNP-based inference of ancestry. J Forensic Sci. 2003;48:771–82. - PubMed
-
- The (current) articles 138a, 151d and 195f of the "wetboek van strafvordering" (the Dutch Criminal Penal Code are available (in Dutch only) online at the following starting page http://wetten.overheid.nl/BWBR0001903/geldigheidsdatum_20-10-2009
-
- Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–99. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

