The GNUMAP algorithm: unbiased probabilistic mapping of oligonucleotides from next-generation sequencing
- PMID: 19861355
- PMCID: PMC6276904
- DOI: 10.1093/bioinformatics/btp614
The GNUMAP algorithm: unbiased probabilistic mapping of oligonucleotides from next-generation sequencing
Abstract
Motivation: The advent of next-generation sequencing technologies has increased the accuracy and quantity of sequence data, opening the door to greater opportunities in genomic research.
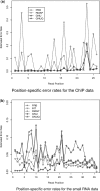
Results: In this article, we present GNUMAP (Genomic Next-generation Universal MAPper), a program capable of overcoming two major obstacles in the mapping of reads from next-generation sequencing runs. First, we have created an algorithm that probabilistically maps reads to repeat regions in the genome on a quantitative basis. Second, we have developed a probabilistic Needleman-Wunsch algorithm which utilizes _prb.txt and _int.txt files produced in the Solexa/Illumina pipeline to improve the mapping accuracy for lower quality reads and increase the amount of usable data produced in a given experiment.
Availability: The source code for the software can be downloaded from http://dna.cs.byu.edu/gnumap.
Figures



References
-
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

