A genomic approach to improve prognosis and predict therapeutic response in chronic lymphocytic leukemia
- PMID: 19861443
- PMCID: PMC2783430
- DOI: 10.1158/1078-0432.CCR-09-1132
A genomic approach to improve prognosis and predict therapeutic response in chronic lymphocytic leukemia
Abstract
Purpose: Chronic lymphocytic leukemia (CLL) is a B-cell malignancy characterized by a variable clinical course. Several parameters have prognostic capabilities but are associated with altered response to therapy in only a small subset of patients.
Experimental design: We used gene expression profiling methods to generate predictors of therapy response and prognosis. Genomic signatures that reflect progressive disease and responses to chemotherapy or chemoimmunotherapy were created using cancer cell lines and patient leukemia cell samples. We validated and applied these three signatures to independent clinical data from four cohorts, representing a total of 301 CLL patients.
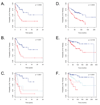
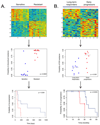
Results: A genomic signature of prognosis created from patient leukemic cell gene expression data coupled with clinical parameters significantly differentiated patients with stable disease from those with progressive disease in the training data set. The progression signature was validated in two independent data sets, showing a capacity to accurately identify patients at risk for progressive disease. In addition, genomic signatures that predict response to chlorambucil or pentostatin, cyclophosphamide, and rituximab were generated and could accurately distinguish responding and nonresponding CLL patients.
Conclusions: Thus, microarray analysis of CLL lymphocytes can be used to refine prognosis and predict response to different therapies. These results have implications for standard and investigational therapeutics in CLL patients.
Figures

References
-
- Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–1775. - PubMed
-
- Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. - PubMed
-
- Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. - PubMed
-
- Hallek M, Wanders L, Ostwald M, et al. Serum beta(2)-microglobulin and serum thymidine kinase are independent predictors of progression-free survival in chronic lymphocytic leukemia and immunocytoma. Leuk Lymphoma. 1996;22:439–447. - PubMed
-
- Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

