Structure of the lethal phage pinhole
- PMID: 19861547
- PMCID: PMC2776468
- DOI: 10.1073/pnas.0907941106
Structure of the lethal phage pinhole
Abstract
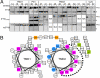
Perhaps the simplest of biological timing systems, bacteriophage holins accumulate during the phage morphogenesis period and then trigger to permeabilize the cytoplasmic membrane with lethal holes; thus, terminating the infection cycle. Canonical holins form very large holes that allow nonspecific release of fully-folded proteins, but a recently discovered class of holins, the pinholins, make much smaller holes, or pinholes, that serve only to depolarize the membrane. Here, we interrogate the structure of the prototype pinholin by negative-stain transmission electron-microscopy, cysteine-accessibility, and chemical cross-linking, as well as by computational approaches. Together, the results suggest that the pinholin forms symmetric heptameric structures with the hydrophilic surface of one transmembrane domain lining the surface of a central channel approximately 15 A in diameter. The structural model also suggests a rationale for the prehole state of the pinholin, the persistence of which defines the duration of the viral latent period, and for the sensitivity of the holin timing system to the energized state of the membrane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Young R, Wang IN. Phage Lysis. In: Calendar R, editor. The Bacteriophages. 2nd Ed. Oxford: Oxford Univ Press; pp. 104–126.
-
- Young R. Bacteriophage holins: Deadly diversity. J Mol Microbiol Biotechnol. 2002;4:21–36. - PubMed
-
- Bläsi U, Young R. Two beginnings for a single purpose: The dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

