Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles
- PMID: 19861549
- PMCID: PMC2776408
- DOI: 10.1073/pnas.0907364106
Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles
Abstract
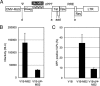
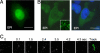
The incorporation of viral genomes into particles has never previously been imaged in live infected cells. Thus, for many viruses it is unknown how the recruitment and packaging of genomes into virions is temporally and spatially related to particle assembly. Here, we devised approaches to simultaneously image HIV-1 genomes, as well as the major HIV-1 structural protein, Gag, to reveal their dynamics and functional interactions during the assembly of individual viral particles. In the absence of Gag, HIV-1 RNA was highly dynamic, moving in and out of the proximity of the plasma membrane. Conversely, in the presence of Gag, RNA molecules docked at the membrane where their lateral movement slowed and then ceased as Gag assembled around them and they became irreversibly anchored. Viral genomes were not retained at the membrane when their packaging signals were mutated, nor when expressed with a Gag mutant that was not myristoylated. In the presence of a Gag mutant that retained membrane- and RNA-binding activities but could not assemble into particles, the viral RNA docked at the membrane but continued to drift laterally and then often dissociated from the membrane. These results, which provide visualization of the recruitment and packaging of genomes into individual virus particles, demonstrate that a small number of Gag molecules recruit viral genomes to the plasma membrane where they nucleate the assembly of complete virions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- D'Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

