Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome
- PMID: 19861685
- PMCID: PMC2829859
- DOI: 10.1182/blood-2009-05-221549
Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome
Abstract
Factor H autoantibodies have been reported in approximately 10% of patients with atypical hemolytic uremic syndrome (aHUS) and are associated with deficiency of factor H-related proteins 1 and 3. In this study we examined the prevalence of factor H autoantibodies in the Newcastle cohort of aHUS patients, determined whether the presence of such autoantibodies is always associated with deficiency of factor H-related proteins 1 and 3, and examined whether such patients have additional susceptibility factors and/or mutations in the genes encoding complement regulator/activators. We screened 142 patients with aHUS and found factor H autoantibodies in 13 individuals (age 1-11 years). The presence of the autoantibodies was confirmed by Western blotting. By using multiplex ligation-dependent probe amplification we measured complement factor H-related (CFHR)1 and CFHR3 copy number. In 10 of the 13 patients there were 0 copies of CFHR1, and in 3 patients there were 2. In 3 of the patients with 0 copies of CFHR1 there was 1 copy of CFHR3, and these individuals exhibited a novel deletion incorporating CFHR1 and CFHR4. In 5 patients mutations were identified: 1 in CFH, 1 in CFI, 1 in CD46, and 2 in C3. The latter observation emphasizes that multiple concurrent factors may be necessary in individual patients for disease manifestation.
Figures

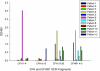

Comment in
-
aHUS: a disorder with many risk factors.Blood. 2010 Jan 14;115(2):158-60. doi: 10.1182/blood-2009-11-252627. Blood. 2010. PMID: 20075170 No abstract available.
References
-
- Kavanagh D, Goodship THJ, Richards A. Atypical haemolytic uraemic syndrome. Br Med Bull. 2006;77–78(1):5–22. - PubMed
-
- Kavanagh D, Kemp EJ, Mayland E, et al. Mutations in complement factor I predispose to development of atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2005;16(7):2150–2155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

