Caenorhabditis is a metazoan host for Legionella
- PMID: 19863556
- PMCID: PMC3120102
- DOI: 10.1111/j.1462-5822.2009.01398.x
Caenorhabditis is a metazoan host for Legionella
Abstract
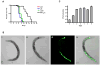
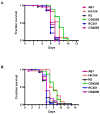
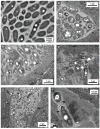
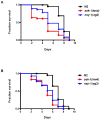
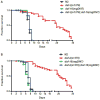
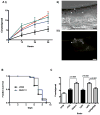
We investigated whether nematodes contribute to the persistence, differentiation and amplification of Legionella species in soil, an emerging source for Legionnaires' disease. Here we show that Legionella spp. colonize the intestinal tracts of Caenorhabditis nematodes leading to worm death. Susceptibility to Legionella is influenced by innate immune responses governed by the p38 mitogen-activated protein kinase and insulin/insulin growth factor-1 receptor signalling pathways. We also show that L. pneumophila colonizes the intestinal tract of nematodes cultivated in soil. To distinguish between transient infection and persistence, plate-fed and soil-extracted nematodes-fed fluorescent strains of L. pneumophila were analysed. Bacteria replicated within the nematode intestinal tract, did not invade surrounding tissue, and were excreted as differentiated forms that were transmitted to offspring. Interestingly, the ultrastructural features of the differentiated bacterial forms were similar to cyst-like forms observed within protozoa, amoeba and mammalian cell lines. While intestinal colonization of L. pneumophila dotA and icmT mutant strains did not alter the survival rate of nematodes in comparison to wild-type strains, nematodes colonized with the dot/icm mutant strains exhibited significantly increased levels of germline apoptosis. Taken together, these studies show that nematodes may serve as natural hosts for these organisms and thereby contribute to their dissemination in the environment and suggest that the remarkable ability of L. pneumophila to subvert host cell signalling and evade mammalian immune responses evolved through the natural selection associated with cycling between protozoan and metazoan hosts.
Figures



References
-
- Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol. 2003;13:47–52. - PubMed
-
- Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–1542. - PubMed
-
- Abu-Zant A, Jones S, Asare R, Suttles J, Price C, Graham J, Kwaik YA. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol. 2007;9:246–264. - PubMed
-
- Anderson GL, Kenney SJ, Millner PD, Beuchat LR, Williams PL. Shedding of foodborne pathogens by Caenorhabditis elegans in compost-amended and unamended soil. Food Microbiol. 2006;23:146–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

