A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps
- PMID: 19863796
- PMCID: PMC2775020
- DOI: 10.1186/1746-4811-5-14
A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps
Abstract
Background: The progress and completion of various plant genome sequencing projects has paved the way for diverse functional genomic studies that involve cloning, modification and subsequent expression of target genes. This requires flexible and efficient procedures for generating binary vectors containing: gene fusions, variants from site-directed mutagenesis, addition of protein tags together with domain swaps and deletions. Furthermore, efficient cloning procedures, ideally high throughput, are essential for pyramiding of multiple gene constructs.
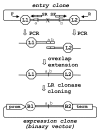
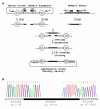
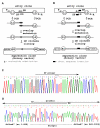
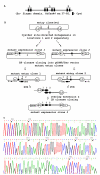
Results: Here, we present a simple, flexible and efficient PCR-fusion/Gateway cloning procedure for construction of binary vectors for a range of gene fusions or variants with single or multiple nucleotide substitutions, short sequence insertions, domain deletions and swaps. Results from selected applications of the procedure which include ORF fusion, introduction of Cys>Ser mutations, insertion of StrepII tag sequence and domain swaps for Arabidopsis secondary cell wall AtCesA genes are demonstrated.
Conclusion: The PCR-fusion/Gateway cloning procedure described provides an elegant, simple and efficient solution for a wide range of diverse and complicated cloning tasks. Through streamlined cloning of sets of gene fusions and modification variants into binary vectors for systematic functional studies of gene families, our method allows for efficient utilization of the growing sequence and expression data.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

