The integrated microbial genomes system: an expanding comparative analysis resource
- PMID: 19864254
- PMCID: PMC2808961
- DOI: 10.1093/nar/gkp887
The integrated microbial genomes system: an expanding comparative analysis resource
Abstract
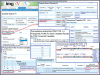
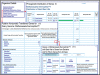
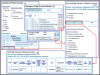
The integrated microbial genomes (IMG) system serves as a community resource for comparative analysis of publicly available genomes in a comprehensive integrated context. IMG contains both draft and complete microbial genomes integrated with other publicly available genomes from all three domains of life, together with a large number of plasmids and viruses. IMG provides tools and viewers for analyzing and reviewing the annotations of genes and genomes in a comparative context. Since its first release in 2005, IMG's data content and analytical capabilities have been constantly expanded through regular releases. Several companion IMG systems have been set up in order to serve domain specific needs, such as expert review of genome annotations. IMG is available at http://img.jgi.doe.gov.
Figures

References
-
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protocols. 2007;2:953–971. - PubMed
-
- Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. - PubMed

