Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses
- PMID: 19864387
- PMCID: PMC2798433
- DOI: 10.1128/JVI.01921-09
Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses
Abstract
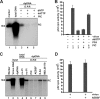
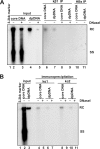
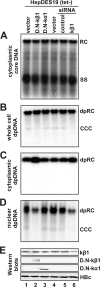
Removal of genome-bound viral DNA polymerase ought to be an essential step in the formation of hepadnavirus covalently closed circular DNA (cccDNA). We previously demonstrated that deproteinized (DP) relaxed circular DNA (rcDNA) of hepatitis B virus (HBV) existed in both the cytoplasm and nuclei of infected cells and the vast majority of cytoplasmic DP rcDNA was associated with DNase I-permeable nucleocapsids. In our efforts to investigate the role of the cytoplasmic DP rcDNA in cccDNA formation, we demonstrated that rcDNA deproteinization could occur in an endogenous DNA polymerase reaction with either virion-derived or intracellular nucleocapsids. As observed in the cytoplasm of virally infected cells, in vitro deproteinization requires the maturation of plus-strand DNA and results in changes in nucleocapsid structure that render the DP rcDNA susceptible to DNase I digestion. Remarkably, we found that the cytoplasmic DP rcDNA-containing nucleocapsids could be selectively immunoprecipitated with an antibody against the carboxyl-terminal peptide of HBV core protein and are associated with cellular nuclear transport receptors karyopherin-alpha and -beta. Moreover, transfection of small interfering RNA targeting karyopherin-beta1 mRNA or expression of a dominant-negative karyopherin-beta1 in a stable cell line supporting HBV replication resulted in the accumulation of DP rcDNA in cytoplasm and reduction of nuclear DP rcDNA and cccDNA. Our results thus favor a hypothesis that completion of plus-strand DNA synthesis triggers the genomic DNA deproteinization and structural changes of nucleocapsids, which leads to the exposure of nuclear localization signals in the C terminus of core protein and mediates the nuclear transportation of DP rcDNA via interaction with karyopherin-alpha and -beta.
Figures




References
-
- Barthelmes, H. U., M. Habermeyer, M. O. Christensen, C. Mielke, H. Interthal, J. J. Pouliot, F. Boege, and D. Marko. 2004. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 279:55618-55625. - PubMed
-
- Chook, Y. M., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703-715. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

