Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones
- PMID: 19864569
- PMCID: PMC2796107
- DOI: 10.1523/JNEUROSCI.3277-09.2009
Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones
Abstract
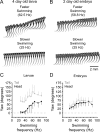
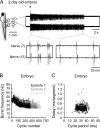
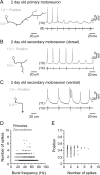
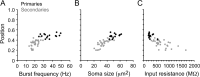
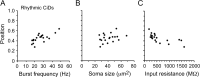
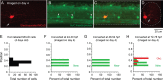
Studies of neuronal networks have revealed few general principles that link patterns of development with later functional roles. While investigating the neural control of movements, we recently discovered a topographic map in the spinal cord of larval zebrafish that relates the position of motoneurons and interneurons to their order of recruitment during swimming. Here, we show that the map reflects an orderly pattern of differentiation of neurons driving different movements. First, we use high-speed filming to show that large-amplitude swimming movements with bending along much of the body appear first, with smaller, regional swimming movements emerging later. Next, using whole-cell patch recordings, we demonstrate that the excitatory circuits that drive large-amplitude, fast swimming movements at larval stages are present and functional early on in embryos. Finally, we systematically assess the orderly emergence of spinal circuits according to swimming speed using transgenic fish expressing the photoconvertible protein Kaede to track neuronal differentiation in vivo. We conclude that a simple principle governs the development of spinal networks in which the neurons driving the fastest, most powerful swimming in larvae develop first with ones that drive increasingly weaker and slower larval movements layered on over time. Because the neurons are arranged by time of differentiation in the spinal cord, the result is a topographic map that represents the speed/strength of movements at which neurons are recruited and the temporal emergence of networks. This pattern may represent a general feature of neuronal network development throughout the brain and spinal cord.
Figures


References
-
- Altman J, Bayer SA. Development of the brain stem in the rat. IV. Thymidine-radiographic study of the time of origin of neurons in the pontine region. J Comp Neurol. 1980;194:905–929. - PubMed
-
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. - PubMed
-
- Beattie CE, Hatta K, Halpern ME, Liu H, Eisen JS, Kimmel CB. Temporal separation in the specification of primary and secondary motoneurons in zebrafish. Dev Biol. 1997;187:171–182. - PubMed
-
- Bekoff A, Trainer W. The development of interlimb co-ordination during swimming in postnatal rats. J Exp Biol. 1979;83:1–11. - PubMed
-
- Berkowitz A. Physiology and morphology of shared and specialized spinal interneurons for locomotion and scratching. J Neurophysiol. 2008;99:2887–2901. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
