Emergent gene order in a model of modular polyketide synthases
- PMID: 19864629
- PMCID: PMC2780807
- DOI: 10.1073/pnas.0902364106
Emergent gene order in a model of modular polyketide synthases
Abstract
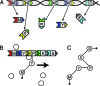
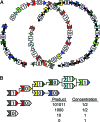
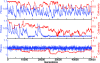
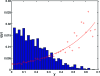
Polyketides are a class of biologically active heteropolymers produced by assembly line-like multiprotein complexes of modular polyketide synthases (PKS). The polyketide product is encoded in the order of the PKS proteins in the assembly line, suggesting that polyketide diversity derives from combinatorial rearrangement of these PKS complexes. Remarkably, the order of PKS genes on the chromosome follows the order of PKS proteins in the assembly line: This fact is commonly referred to as "collinearity". Here we propose an evolutionary origin for collinearity and demonstrate the mechanism by using a computational model of PKS evolution in a population. Assuming continuous evolutionary pressure for novel polyketides, and that new polyketide pathways are formed by horizontal transfer/recombination of PKS-encoding DNA, we demonstrate the existence of a broad range of parameters for which collinearity emerges spontaneously. Collinearity confers no fitness advantage in our model; it is established and maintained through a "secondary selection" mechanism, as a trait which increases the probability of forming long, novel PKS complexes through recombination. Consequently, collinearity hitchhikes on the successful genotypes which periodically sweep through the evolving population. In addition to computer simulation of a simplified model of PKS evolution, we provide a mathematical framework describing the secondary selection mechanism, which generalizes beyond the context of the present model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gokhale, et al. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat Prod Rep. 2007;24:267–277. - PubMed
-
- Firn RD, Jones CG. Natural products—A simple model to explain chemical diversity. Nat Prod Rep. 2003;20:382–391. - PubMed
-
- Staunton J, Weissman K. Polyketide biosynthesis: A millennium review. Nat Prod Rep. 2001;18:380–416. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

